+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0741 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryoEM 3D reconstruction of IGF1R in complex with IGF1 | |||||||||
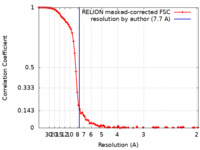
 Map data Map data | postprocess resolution at 7.68A by Relion3. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.7 Å | |||||||||
 Authors Authors | Wang T / Sun J | |||||||||
 Citation Citation |  Journal: Structure / Year: 2020 Journal: Structure / Year: 2020Title: Visualization of Ligand-Bound Ectodomain Assembly in the Full-Length Human IGF-1 Receptor by Cryo-EM Single-Particle Analysis. Authors: Xi Zhang / Daqi Yu / Jingchuan Sun / Yujie Wu / Junyuan Gong / Xuemei Li / Li Liu / Shan Liu / Jianbo Liu / Yulan Wu / Dongyang Li / Yinping Ma / Xu Han / Yanan Zhu / Zhaolong Wu / Yihua ...Authors: Xi Zhang / Daqi Yu / Jingchuan Sun / Yujie Wu / Junyuan Gong / Xuemei Li / Li Liu / Shan Liu / Jianbo Liu / Yulan Wu / Dongyang Li / Yinping Ma / Xu Han / Yanan Zhu / Zhaolong Wu / Yihua Wang / Qi Ouyang / Tao Wang /  Abstract: Tyrosine kinase receptor of insulin-like growth factor 1 receptor (IGF-1R) and insulin receptor (IR) bind to hormones, such as insulin, IGF-1, and IGF-2, and transduces the signals across the cell ...Tyrosine kinase receptor of insulin-like growth factor 1 receptor (IGF-1R) and insulin receptor (IR) bind to hormones, such as insulin, IGF-1, and IGF-2, and transduces the signals across the cell membrane. However, the complete structure of the receptor and the signal transduction mechanism remains unclear. Here, we report the cryo-EM structure of the ligand-bound ectodomain in the full-length human IGF-1R. We reconstructed the IGF-1R/insulin complex at 4.7 Å and the IGF-1R/IGF-1 complex at 7.7 Å. Our structures reveal that only one insulin or one IGF-1 molecule binds to and activates the full-length human IGF-1R receptor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0741.map.gz emd_0741.map.gz | 2.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0741-v30.xml emd-0741-v30.xml emd-0741.xml emd-0741.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0741_fsc.xml emd_0741_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_0741.png emd_0741.png | 67.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0741 http://ftp.pdbj.org/pub/emdb/structures/EMD-0741 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0741 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0741 | HTTPS FTP |
-Validation report
| Summary document |  emd_0741_validation.pdf.gz emd_0741_validation.pdf.gz | 78.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0741_full_validation.pdf.gz emd_0741_full_validation.pdf.gz | 77.3 KB | Display | |
| Data in XML |  emd_0741_validation.xml.gz emd_0741_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0741 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0741 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0741 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0741 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0741.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0741.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocess resolution at 7.68A by Relion3. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.99 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : IGF1R
| Entire | Name: IGF1R |
|---|---|
| Components |
|
-Supramolecule #1: IGF1R
| Supramolecule | Name: IGF1R / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 300 kDa/nm |
-Macromolecule #1: IGF1 receptor
| Macromolecule | Name: IGF1 receptor / type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKSGSGGGSP TSLWGLLFLS AALSLWPTSG EICGPGIDIR NDYQQLKRLE NCTVIEGYLH ILLISKAEDY RSYRFPKLT VITEYLLLFR VAGLESLGDL FPNLTVIRGW KLFYNYALVI FEMTNLKDIG LYNLRNITRG A IRIEKNAD LCYLSTVDWS LILDAVSNNY ...String: MKSGSGGGSP TSLWGLLFLS AALSLWPTSG EICGPGIDIR NDYQQLKRLE NCTVIEGYLH ILLISKAEDY RSYRFPKLT VITEYLLLFR VAGLESLGDL FPNLTVIRGW KLFYNYALVI FEMTNLKDIG LYNLRNITRG A IRIEKNAD LCYLSTVDWS LILDAVSNNY IVGNKPPKEC GDLCPGTMEE KPMCEKTTIN NEYNYRCWTT NR CQKMCPS TCGKRACTEN NECCHPECLG SCSAPDNDTA CVACRHYYYA GVCVPACPPN TYRFEGWRCV DRD FCANIL SAESSDSEGF VIHDGECMQE CPSGFIRNGS QSMYCIPCEG PCPKVCEEEK KTKTIDSVTS AQML QGCTI FKGNLLINIR RGNNIASELE NFMGLIEVVT GYVKIRHSHA LVSLSFLKNL RLILGEEQLE GNYSF YVLD NQNLQQLWDW DHRNLTIKAG KMYFAFNPKL CVSEIYRMEE VTGTKGRQSK GDINTRNNGE RASCES DVL HFTSTTTSKN RIIITWHRYR PPDYRDLISF TVYYKEAPFK NVTEYDGQDA CGSNSWNMVD VDLPPNK DV EPGILLHGLK PWTQYAVYVK AVTLTMVEND HIRGAKSEIL YIRTNASVPS IPLDVLSASN SSSQLIVK W NPPSLPNGNL SYYIVRWQRQ PQDGYLYRHN YCSKDKIPIR KYADGTIDIE EVTENPKTEV CGGEKGPCC ACPKTEAEKQ AEKEEAEYRK VFENFLHNSI FVPRPERKRR DVMQVANTTM SSRSRNTTAA DTYNITDPEE LETEYPFFE SRVDNKERTV ISNLRPFTLY RIDIHSCNHE AEKLGCSASN FVFARTMPAE GADDIPGPVT W EPRPENSI FLKWPEPENP NGLILMYEIK YGSQVEDQRE CVSRQEYRKY GGAKLNRLNP GNYTARIQAT SL SGNGSWT DPVFFYVQAK TGYENFIHLI IALPVAVLLI VGGLVIMLYV FHRKRNNSRL GNGVLYASVN PEY FSAADV YVPDEWEVAR EKITMSRELG QGSFGMVYEG VAKGVVKDEP ETRVAIKTVN EAASMRERIE FLNE ASVMK EFNCHHVVRL LGVVSQGQPT LVIMELMTRG DLKSYLRSLR PEMENNPVLA PPSLSKMIQM AGEIA DGMA YLNANKFVHR DLAARNCMVA EDFTVKIGDF GMTRDIYETD YYRKGGKGLL PVRWMSPESL KDGVFT TYS DVWSFGVVLW EIATLAEQPY QGLSNEQVLR FVMEGGLLDK PDNCPDMLFE LMRMCWQYNP KMRPSFL EI ISSIKEEMEP GFREVSFYYS EENKLPEPEE LDLEPENMES VPLDPSASSS SLPLPDRHSG HKAENGPG P GVLVLRASFD ERQPYAHMNG GRKNERALPL PQSSTC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7 / Details: PBS buffer with 0.05% (w/v) DDM |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 50.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 11 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)