[English] 日本語
 Yorodumi
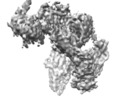
Yorodumi- EMDB-9838: Cryo-EM structure of the full-length human IGF-1R in complex with... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9838 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the full-length human IGF-1R in complex with insulin | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | human type 1 insulin-like growth factor receptor / insulin / SIGNALING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationinsulin-like growth factor receptor activity / protein kinase complex / insulin-like growth factor binding / Signaling by Type 1 Insulin-like Growth Factor 1 Receptor (IGF1R) / IRS-related events triggered by IGF1R / protein transporter activity / transcytosis / insulin receptor complex / positive regulation of protein-containing complex disassembly / insulin-like growth factor I binding ...insulin-like growth factor receptor activity / protein kinase complex / insulin-like growth factor binding / Signaling by Type 1 Insulin-like Growth Factor 1 Receptor (IGF1R) / IRS-related events triggered by IGF1R / protein transporter activity / transcytosis / insulin receptor complex / positive regulation of protein-containing complex disassembly / insulin-like growth factor I binding / insulin receptor activity / alphav-beta3 integrin-IGF-1-IGF1R complex / dendritic spine maintenance / peptidyl-tyrosine autophosphorylation / regulation of JNK cascade / insulin binding / negative regulation of glycogen catabolic process / positive regulation of nitric oxide mediated signal transduction / negative regulation of fatty acid metabolic process / negative regulation of feeding behavior / Signaling by Insulin receptor / IRS activation / regulation of protein secretion / Insulin processing / positive regulation of peptide hormone secretion / positive regulation of respiratory burst / negative regulation of acute inflammatory response / Regulation of gene expression in beta cells / amyloid-beta clearance / alpha-beta T cell activation / Respiratory syncytial virus (RSV) attachment and entry / insulin receptor substrate binding / positive regulation of dendritic spine maintenance / Synthesis, secretion, and deacylation of Ghrelin / negative regulation of protein secretion / negative regulation of gluconeogenesis / positive regulation of glycogen biosynthetic process / fatty acid homeostasis / Signal attenuation / positive regulation of insulin receptor signaling pathway / negative regulation of respiratory burst involved in inflammatory response / FOXO-mediated transcription of oxidative stress, metabolic and neuronal genes / SHC-related events triggered by IGF1R / negative regulation of lipid catabolic process / positive regulation of lipid biosynthetic process / negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / regulation of protein localization to plasma membrane / phosphatidylinositol 3-kinase binding / nitric oxide-cGMP-mediated signaling / transport vesicle / COPI-mediated anterograde transport / positive regulation of nitric-oxide synthase activity / negative regulation of MAPK cascade / Insulin receptor recycling / negative regulation of reactive oxygen species biosynthetic process / positive regulation of brown fat cell differentiation / insulin-like growth factor receptor binding / NPAS4 regulates expression of target genes / neuron projection maintenance / endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of mitotic nuclear division / insulin-like growth factor receptor signaling pathway / Insulin receptor signalling cascade / positive regulation of glycolytic process / positive regulation of cytokine production / endosome lumen / positive regulation of long-term synaptic potentiation / acute-phase response / positive regulation of D-glucose import across plasma membrane / positive regulation of protein secretion / insulin receptor binding / phosphatidylinositol 3-kinase/protein kinase B signal transduction / positive regulation of cell differentiation / Regulation of insulin secretion / wound healing / cellular response to glucose stimulus / receptor protein-tyrosine kinase / positive regulation of neuron projection development / hormone activity / regulation of synaptic plasticity / negative regulation of protein catabolic process / positive regulation of protein localization to nucleus / Golgi lumen / vasodilation / cognition / glucose metabolic process / cellular response to amyloid-beta / insulin receptor signaling pathway / cell-cell signaling / glucose homeostasis / regulation of protein localization / positive regulation of cold-induced thermogenesis / protein autophosphorylation / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / positive regulation of cell growth / protease binding / protein tyrosine kinase activity / secretory granule lumen / Extra-nuclear estrogen signaling / positive regulation of canonical NF-kappaB signal transduction Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.0 Å | ||||||||||||
 Authors Authors | Zhang X / Yu D | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Structure / Year: 2020 Journal: Structure / Year: 2020Title: Visualization of Ligand-Bound Ectodomain Assembly in the Full-Length Human IGF-1 Receptor by Cryo-EM Single-Particle Analysis. Authors: Xi Zhang / Daqi Yu / Jingchuan Sun / Yujie Wu / Junyuan Gong / Xuemei Li / Li Liu / Shan Liu / Jianbo Liu / Yulan Wu / Dongyang Li / Yinping Ma / Xu Han / Yanan Zhu / Zhaolong Wu / Yihua ...Authors: Xi Zhang / Daqi Yu / Jingchuan Sun / Yujie Wu / Junyuan Gong / Xuemei Li / Li Liu / Shan Liu / Jianbo Liu / Yulan Wu / Dongyang Li / Yinping Ma / Xu Han / Yanan Zhu / Zhaolong Wu / Yihua Wang / Qi Ouyang / Tao Wang /  Abstract: Tyrosine kinase receptor of insulin-like growth factor 1 receptor (IGF-1R) and insulin receptor (IR) bind to hormones, such as insulin, IGF-1, and IGF-2, and transduces the signals across the cell ...Tyrosine kinase receptor of insulin-like growth factor 1 receptor (IGF-1R) and insulin receptor (IR) bind to hormones, such as insulin, IGF-1, and IGF-2, and transduces the signals across the cell membrane. However, the complete structure of the receptor and the signal transduction mechanism remains unclear. Here, we report the cryo-EM structure of the ligand-bound ectodomain in the full-length human IGF-1R. We reconstructed the IGF-1R/insulin complex at 4.7 Å and the IGF-1R/IGF-1 complex at 7.7 Å. Our structures reveal that only one insulin or one IGF-1 molecule binds to and activates the full-length human IGF-1R receptor. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9838.map.gz emd_9838.map.gz | 1.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9838-v30.xml emd-9838-v30.xml emd-9838.xml emd-9838.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9838_fsc.xml emd_9838_fsc.xml | 5.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_9838.png emd_9838.png | 160.3 KB | ||
| Filedesc metadata |  emd-9838.cif.gz emd-9838.cif.gz | 8.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9838 http://ftp.pdbj.org/pub/emdb/structures/EMD-9838 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9838 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9838 | HTTPS FTP |
-Related structure data
| Related structure data |  6jk8MC  0741C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9838.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9838.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.37 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : complex of full-length human type 1 insulin-like growth factor re...
| Entire | Name: complex of full-length human type 1 insulin-like growth factor receptor with insulin |
|---|---|
| Components |
|
-Supramolecule #1: complex of full-length human type 1 insulin-like growth factor re...
| Supramolecule | Name: complex of full-length human type 1 insulin-like growth factor receptor with insulin type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 310 KDa |
-Macromolecule #1: Insulin-like growth factor 1 receptor
| Macromolecule | Name: Insulin-like growth factor 1 receptor / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: receptor protein-tyrosine kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 154.964469 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKSGSGGGSP TSLWGLLFLS AALSLWPTSG EICGPGIDIR NDYQQLKRLE NCTVIEGYLH ILLISKAEDY RSYRFPKLTV ITEYLLLFR VAGLESLGDL FPNLTVIRGW KLFYNYALVI FEMTNLKDIG LYNLRNITRG AIRIEKNADL CYLSTVDWSL I LDAVSNNY ...String: MKSGSGGGSP TSLWGLLFLS AALSLWPTSG EICGPGIDIR NDYQQLKRLE NCTVIEGYLH ILLISKAEDY RSYRFPKLTV ITEYLLLFR VAGLESLGDL FPNLTVIRGW KLFYNYALVI FEMTNLKDIG LYNLRNITRG AIRIEKNADL CYLSTVDWSL I LDAVSNNY IVGNKPPKEC GDLCPGTMEE KPMCEKTTIN NEYNYRCWTT NRCQKMCPST CGKRACTENN ECCHPECLGS CS APDNDTA CVACRHYYYA GVCVPACPPN TYRFEGWRCV DRDFCANILS AESSDSEGFV IHDGECMQEC PSGFIRNGSQ SMY CIPCEG PCPKVCEEEK KTKTIDSVTS AQMLQGCTIF KGNLLINIRR GNNIASELEN FMGLIEVVTG YVKIRHSHAL VSLS FLKNL RLILGEEQLE GNYSFYVLDN QNLQQLWDWD HRNLTIKAGK MYFAFNPKLC VSEIYRMEEV TGTKGRQSKG DINTR NNGE RASCESDVLH FTSTTTSKNR IIITWHRYRP PDYRDLISFT VYYKEAPFKN VTEYDGQDAC GSNSWNMVDV DLPPNK DVE PGILLHGLKP WTQYAVYVKA VTLTMVENDH IRGAKSEILY IRTNASVPSI PLDVLSASNS SSQLIVKWNP PSLPNGN LS YYIVRWQRQP QDGYLYRHNY CSKDKIPIRK YADGTIDIEE VTENPKTEVC GGEKGPCCAC PKTEAEKQAE KEEAEYRK V FENFLHNSIF VPRPERKRRD VMQVANTTMS SRSRNTTAAD TYNITDPEEL ETEYPFFESR VDNKERTVIS NLRPFTLYR IDIHSCNHEA EKLGCSASNF VFARTMPAEG ADDIPGPVTW EPRPENSIFL KWPEPENPNG LILMYEIKYG SQVEDQRECV SRQEYRKYG GAKLNRLNPG NYTARIQATS LSGNGSWTDP VFFYVQAKTG YENFIHLIIA LPVAVLLIVG GLVIMLYVFH R KRNNSRLG NGVLYASVNP EYFSAADVYV PDEWEVAREK ITMSRELGQG SFGMVYEGVA KGVVKDEPET RVAIKTVNEA AS MRERIEF LNEASVMKEF NCHHVVRLLG VVSQGQPTLV IMELMTRGDL KSYLRSLRPE MENNPVLAPP SLSKMIQMAG EIA DGMAYL NANKFVHRDL AARNCMVAED FTVKIGDFGM TRDIYETDYY RKGGKGLLPV RWMSPESLKD GVFTTYSDVW SFGV VLWEI ATLAEQPYQG LSNEQVLRFV MEGGLLDKPD NCPDMLFELM RMCWQYNPKM RPSFLEIISS IKEEMEPGFR EVSFY YSEE NKLPEPEELD LEPENMESVP LDPSASSSSL PLPDRHSGHK AENGPGPGVL VLRASFDERQ PYAHMNGGRK NERALP LPQ SSTC UniProtKB: Insulin-like growth factor 1 receptor |
-Macromolecule #2: Insulin
| Macromolecule | Name: Insulin / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.989862 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MALWMRLLPL LALLALWGPD PAAAFVNQHL CGSHLVEALY LVCGERGFFY TPKTRREAED LQVGQVELGG GPGAGSLQPL ALEGSLQKR GIVEQCCTSI CSLYQLENYC N UniProtKB: Insulin |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: PBS with detergent |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
| Details | human type 1 insulin-like growth factor receptor saturated with human insulin |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 36496 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)























