+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31982 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MT2-remalteon-Gi complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | G protein coupled receptor / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of circadian sleep/wake cycle, non-REM sleep / melatonin receptor activity / regulation of neuronal action potential / negative regulation of transmission of nerve impulse / positive regulation of transmission of nerve impulse / Class A/1 (Rhodopsin-like receptors) / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor ...positive regulation of circadian sleep/wake cycle, non-REM sleep / melatonin receptor activity / regulation of neuronal action potential / negative regulation of transmission of nerve impulse / positive regulation of transmission of nerve impulse / Class A/1 (Rhodopsin-like receptors) / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / camera-type eye development / Ca2+ pathway / G alpha (z) signalling events / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / G alpha (q) signalling events / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (i) signalling events / alkylglycerophosphoethanolamine phosphodiesterase activity / photoreceptor outer segment membrane / negative regulation of vasoconstriction / negative regulation of cGMP-mediated signaling / spectrin binding / negative regulation of cytosolic calcium ion concentration / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / photoreceptor outer segment / T cell migration / Adenylate cyclase inhibitory pathway / negative regulation of insulin secretion / positive regulation of protein localization to cell cortex / regulation of cAMP-mediated signaling / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / cardiac muscle cell apoptotic process / regulation of mitotic spindle organization / cellular response to forskolin / regulation of insulin secretion / photoreceptor inner segment / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / Regulation of insulin secretion / G protein-coupled receptor binding / G protein-coupled receptor activity / G-protein beta/gamma-subunit complex binding / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / response to peptide hormone / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / G alpha (z) signalling events / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / GPER1 signaling / GDP binding / heterotrimeric G-protein complex / signaling receptor complex adaptor activity / GTPase binding / glucose homeostasis / retina development in camera-type eye / phospholipase C-activating G protein-coupled receptor signaling pathway / positive regulation of cytosolic calcium ion concentration / cell cortex / cellular response to hypoxia / cell body / midbody / G alpha (i) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / G alpha (s) signalling events / chemical synaptic transmission / negative regulation of neuron apoptotic process / cell population proliferation / Extra-nuclear estrogen signaling / G protein-coupled receptor signaling pathway / cell division / lysosomal membrane / GTPase activity / centrosome / dendrite / synapse / protein-containing complex binding / nucleolus / GTP binding / magnesium ion binding / extracellular exosome / nucleoplasm / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.46 Å | |||||||||
 Authors Authors | Wang QG / Lu QY | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis of the ligand binding and signaling mechanism of melatonin receptors. Authors: Qinggong Wang / Qiuyuan Lu / Qiong Guo / Maikun Teng / Qingguo Gong / Xu Li / Yang Du / Zheng Liu / Yuyong Tao /  Abstract: Melatonin receptors (MT and MT in humans) are family A G protein-coupled receptors that respond to the neurohormone melatonin to regulate circadian rhythm and sleep. Numerous efforts have been made ...Melatonin receptors (MT and MT in humans) are family A G protein-coupled receptors that respond to the neurohormone melatonin to regulate circadian rhythm and sleep. Numerous efforts have been made to develop drugs targeting melatonin receptors for the treatment of insomnia, circadian rhythm disorder, and cancer. However, designing subtype-selective melatonergic drugs remains challenging. Here, we report the cryo-EM structures of the MT-G signaling complex with 2-iodomelatonin and ramelteon and the MT-G signaling complex with ramelteon. These structures, together with the reported functional data, reveal that although MT and MT possess highly similar orthosteric ligand-binding pockets, they also display distinctive features that could be targeted to design subtype-selective drugs. The unique structural motifs in MT and MT mediate structural rearrangements with a particularly wide opening on the cytoplasmic side. G is engaged in the receptor core shared by MT and MT and presents a conformation deviating from those in other G complexes. Together, our results provide new clues for designing melatonergic drugs and further insights into understanding the G protein coupling mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31982.map.gz emd_31982.map.gz | 33 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31982-v30.xml emd-31982-v30.xml emd-31982.xml emd-31982.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31982.png emd_31982.png | 43.8 KB | ||
| Filedesc metadata |  emd-31982.cif.gz emd-31982.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31982 http://ftp.pdbj.org/pub/emdb/structures/EMD-31982 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31982 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31982 | HTTPS FTP |
-Validation report
| Summary document |  emd_31982_validation.pdf.gz emd_31982_validation.pdf.gz | 510 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31982_full_validation.pdf.gz emd_31982_full_validation.pdf.gz | 509.6 KB | Display | |
| Data in XML |  emd_31982_validation.xml.gz emd_31982_validation.xml.gz | 6.3 KB | Display | |
| Data in CIF |  emd_31982_validation.cif.gz emd_31982_validation.cif.gz | 7.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31982 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31982 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31982 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31982 | HTTPS FTP |
-Related structure data
| Related structure data |  7vh0MC  7vgyC  7vgzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31982.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31982.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : melatonin receptor 2 with G peotein complex
| Entire | Name: melatonin receptor 2 with G peotein complex |
|---|---|
| Components |
|
-Supramolecule #1: melatonin receptor 2 with G peotein complex
| Supramolecule | Name: melatonin receptor 2 with G peotein complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Melatonin receptor type 1B
| Macromolecule | Name: Melatonin receptor type 1B / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.308285 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSENGSFANC CEAGGWAVRP GWSGAGSARP SRTPRPPWVA PALSAVLIVT TAVDVVGNLL VILSVLRNRK LRNAGNLFLV SLALADLVV AFYPYPLILV AIFYDGWAFG EEHCKASAFV MGLSVIGSVW NITAIAINRY LYICHSMAYH RIYRRWHTPL H ICLIWLLT ...String: MSENGSFANC CEAGGWAVRP GWSGAGSARP SRTPRPPWVA PALSAVLIVT TAVDVVGNLL VILSVLRNRK LRNAGNLFLV SLALADLVV AFYPYPLILV AIFYDGWAFG EEHCKASAFV MGLSVIGSVW NITAIAINRY LYICHSMAYH RIYRRWHTPL H ICLIWLLT VVALLPNFFV GSLEYDPRIY SCTFIQTAST QYTAAVVVIH FLLPIAVVSF CYLRIWVLVL QARRKAKPES RL CLKPSDL RSFLTMFVVF VIFAICWAPL NCIGLAVAIN PQEMAPQIPE GLFVTSYLLA YFNSCLNAIV YGLLNQNFRR EYK RILLAL WNPRHCIQDA SKGSHAEGLQ SPAPPIIGVQ HQADAL UniProtKB: Melatonin receptor type 1B |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.340887 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: GGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGGQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCA TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.021648 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHHHMG SLLQSELDEL RQEAEQLKNQ IRDARKACAD ATLSQITNNI DPVGRIQMRT RRTLRGHLAK IYAMHWGTDS RLLVSASQD GKLIIWDSYT TNKVHAIPLR SSWVMTCAYA PSGNYVACGG LDNICSIYNL KTRQGNVRVS RELAGHTGYL S CCRFLDDN ...String: HHHHHHHHMG SLLQSELDEL RQEAEQLKNQ IRDARKACAD ATLSQITNNI DPVGRIQMRT RRTLRGHLAK IYAMHWGTDS RLLVSASQD GKLIIWDSYT TNKVHAIPLR SSWVMTCAYA PSGNYVACGG LDNICSIYNL KTRQGNVRVS RELAGHTGYL S CCRFLDDN QIVTSSGDTT CALWDIETGQ QTTTFTGHTG DVMSLSLAPD TRLFVSGACD ASAKLWDVRE GMCRQTFTGH ES DINAICF FPDGNAFATG SDDATCRLFD LRADQELMTY SHDNIICGIT SVSFSKSGRL LLAGYDDFNC NVWDALKADR AGV LAGHDN RVSCLGVTDD GMAVATGSWD SFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.767035 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ASNNTASIAQ ARKLVQQLKM EANIDRIKVS KAAADLMAYC EAHAKEDPLL TPVPASQNPF REKKFFCDVQ LVESGGGLVQ PGGSRKLSC SASGFAFSSF GMHWVRQAPE KGLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA M YYCVRSIY ...String: ASNNTASIAQ ARKLVQQLKM EANIDRIKVS KAAADLMAYC EAHAKEDPLL TPVPASQNPF REKKFFCDVQ LVESGGGLVQ PGGSRKLSC SASGFAFSSF GMHWVRQAPE KGLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA M YYCVRSIY YYGSSPFDFW GQGTTLTVSS GGGGSGGGGS GGGGSDIVMT QATSSVPVTP GESVSISCRS SKSLLHSNGN TY LYWFLQR PGQSPQLLIY RMSNLASGVP ERFSGSGSGT AFTLTISRLE AEDVGVYYCM QHLEYPLTFG AGTKLELKGS LEV LFQ |
-Macromolecule #5: N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}pr...
| Macromolecule | Name: N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}propanamide type: ligand / ID: 5 / Number of copies: 1 / Formula: JEV |
|---|---|
| Molecular weight | Theoretical: 259.343 Da |
| Chemical component information | 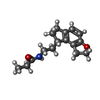 ChemComp-JEV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.46 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 335170 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller




































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















