+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31980 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Melatonin receptor1-2-Iodomelatonin-Gicomplex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | G protein coupled receptor / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmelatonin receptor activity / Class A/1 (Rhodopsin-like receptors) / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding ...melatonin receptor activity / Class A/1 (Rhodopsin-like receptors) / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G alpha (z) signalling events / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / G alpha (q) signalling events / mating behavior / G alpha (i) signalling events / Thrombin signalling through proteinase activated receptors (PARs) / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G alpha (12/13) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Ca2+ pathway / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (z) signalling events / Extra-nuclear estrogen signaling / G alpha (s) signalling events / photoreceptor outer segment membrane / G alpha (q) signalling events / spectrin binding / G alpha (i) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Vasopressin regulates renal water homeostasis via Aquaporins / alkylglycerophosphoethanolamine phosphodiesterase activity / hormone binding / photoreceptor outer segment / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / response to prostaglandin E / D2 dopamine receptor binding / cardiac muscle cell apoptotic process / photoreceptor inner segment / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / regulation of mitotic spindle organization / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / circadian rhythm / negative regulation of insulin secretion / G protein-coupled receptor binding / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to peptide hormone / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / ADP signalling through P2Y purinoceptor 12 / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / G alpha (z) signalling events / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / sensory perception of taste / signaling receptor complex adaptor activity / positive regulation of cytosolic calcium ion concentration / retina development in camera-type eye / cell body / G protein activity / GTPase binding / midbody / cell cortex / G alpha (i) signalling events Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /    | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Wang QG / Lu QY | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis of the ligand binding and signaling mechanism of melatonin receptors. Authors: Qinggong Wang / Qiuyuan Lu / Qiong Guo / Maikun Teng / Qingguo Gong / Xu Li / Yang Du / Zheng Liu / Yuyong Tao /  Abstract: Melatonin receptors (MT and MT in humans) are family A G protein-coupled receptors that respond to the neurohormone melatonin to regulate circadian rhythm and sleep. Numerous efforts have been made ...Melatonin receptors (MT and MT in humans) are family A G protein-coupled receptors that respond to the neurohormone melatonin to regulate circadian rhythm and sleep. Numerous efforts have been made to develop drugs targeting melatonin receptors for the treatment of insomnia, circadian rhythm disorder, and cancer. However, designing subtype-selective melatonergic drugs remains challenging. Here, we report the cryo-EM structures of the MT-G signaling complex with 2-iodomelatonin and ramelteon and the MT-G signaling complex with ramelteon. These structures, together with the reported functional data, reveal that although MT and MT possess highly similar orthosteric ligand-binding pockets, they also display distinctive features that could be targeted to design subtype-selective drugs. The unique structural motifs in MT and MT mediate structural rearrangements with a particularly wide opening on the cytoplasmic side. G is engaged in the receptor core shared by MT and MT and presents a conformation deviating from those in other G complexes. Together, our results provide new clues for designing melatonergic drugs and further insights into understanding the G protein coupling mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31980.map.gz emd_31980.map.gz | 117.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31980-v30.xml emd-31980-v30.xml emd-31980.xml emd-31980.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31980.png emd_31980.png | 42 KB | ||
| Filedesc metadata |  emd-31980.cif.gz emd-31980.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31980 http://ftp.pdbj.org/pub/emdb/structures/EMD-31980 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31980 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31980 | HTTPS FTP |
-Related structure data
| Related structure data |  7vgyMC  7vgzC  7vh0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31980.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31980.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : G protein coupled to melatonin receptor
| Entire | Name: G protein coupled to melatonin receptor |
|---|---|
| Components |
|
-Supramolecule #1: G protein coupled to melatonin receptor
| Supramolecule | Name: G protein coupled to melatonin receptor / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Melatonin receptor type 1A
| Macromolecule | Name: Melatonin receptor type 1A / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.408594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQGNGSALPN ASQPVLRGDG ARPSWLASAL ACVLIFTIVV DILGNLLVIL SVYRNKKLRN AGNIFVVSLA VADLVVAIYP YPLVLMSIF NNGWNLGYLH CQVSGFLMGL SVIGSIFNIT GIAINRYCYI CHSLKYDKLY SSKNSLCYVL LIWLLTLAAV L PNLRAGTL ...String: MQGNGSALPN ASQPVLRGDG ARPSWLASAL ACVLIFTIVV DILGNLLVIL SVYRNKKLRN AGNIFVVSLA VADLVVAIYP YPLVLMSIF NNGWNLGYLH CQVSGFLMGL SVIGSIFNIT GIAINRYCYI CHSLKYDKLY SSKNSLCYVL LIWLLTLAAV L PNLRAGTL QYDPRIYSCT FAQSVSSAYT IAVVVFHFLV PMIIVIFCYL RIWILVLQVR QRVKPDRKPK LKPQDFRNFV TM FVVFVLF AICWAPLNFI GLAVASDPAS MVPRIPEWLF VASYYMAYFN SCLNAIIYGL LNQNFRKEYR RIIVSLCTAR VFF VDSSND VADRVKWKPS PLMTNNNVVK VDSV UniProtKB: Melatonin receptor type 1A |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.283836 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GCTLSAEDKA AVERSKMIDR NLREDGEKAA REVKLLLLGA GESGKSTIVK QMKIIHEAGY SEEECKQYKA VVYSNTIQSI IAIIRAMGR LKIDFGDSAR ADDARQLFVL AGAAEEGFMT AELAGVIKRL WKDSGVQACF NRSREYQLND SAAYYLNDLD R IAQPNYIP ...String: GCTLSAEDKA AVERSKMIDR NLREDGEKAA REVKLLLLGA GESGKSTIVK QMKIIHEAGY SEEECKQYKA VVYSNTIQSI IAIIRAMGR LKIDFGDSAR ADDARQLFVL AGAAEEGFMT AELAGVIKRL WKDSGVQACF NRSREYQLND SAAYYLNDLD R IAQPNYIP TQQDVLRTRV KTTGIVETHF TFKDLHFKMF DVGGQRSERK KWIHCFEGVT AIIFCVALSD YDLVLAEDEE MN RMHESMK LFDSICNNKW FTDTSIILFL NKKDLFEEKI KKSPLTICYP EYAGSNTYEE AAAYIQCQFE DLNKRKDTKE IYT HFTCAT DTKNVQFVFD AVTDVIIKNN LKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.728152 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPGSSGSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: GPGSSGSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.432554 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ASNNTASIAQ ARKLVEQLKM EANIDRIKVS KAAADLMAYC EAHAKEDPLL TPVPASENPF REKKFFC UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.340482 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KGSLEVLFQ |
-Macromolecule #6: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 6 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #7: N-[2-(2-iodo-5-methoxy-1H-indol-3-yl)ethyl]acetamide
| Macromolecule | Name: N-[2-(2-iodo-5-methoxy-1H-indol-3-yl)ethyl]acetamide / type: ligand / ID: 7 / Number of copies: 1 / Formula: ML2 |
|---|---|
| Molecular weight | Theoretical: 358.175 Da |
| Chemical component information | 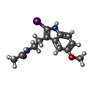 ChemComp-ML2: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















