+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-23940 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of the human SSU processome, state post-A1 - raw maps | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報perforant pathway to dendrate granule cell synapse / mRNA N-acetyltransferase activity / preribosome / regulation of translation at postsynapse / U6 snRNA 2'-O-ribose methyltransferase activity / oocyte growth / nucleologenesis / rRNA cytidine N-acetyltransferase activity / leucine zipper domain binding / snoRNA localization ...perforant pathway to dendrate granule cell synapse / mRNA N-acetyltransferase activity / preribosome / regulation of translation at postsynapse / U6 snRNA 2'-O-ribose methyltransferase activity / oocyte growth / nucleologenesis / rRNA cytidine N-acetyltransferase activity / leucine zipper domain binding / snoRNA localization / granular component / rRNA acetylation involved in maturation of SSU-rRNA / tRNA N-acetyltransferase activity / t-UTP complex / tRNA acetylation / CURI complex / UTP-C complex / Pwp2p-containing subcomplex of 90S preribosome / regulation of stem cell population maintenance / U4atac snRNP / histone H2AQ104 methyltransferase activity / negative regulation of amyloid precursor protein biosynthetic process / Mpp10 complex / U4atac snRNA binding / rRNA (pseudouridine) methyltransferase activity / rRNA modification / pre-snoRNP complex / endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / box C/D sno(s)RNA binding / box C/D sno(s)RNA 3'-end processing / dense fibrillar component / endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / regulation of centrosome duplication / rRNA methyltransferase activity / histone methyltransferase binding / positive regulation of rRNA processing / N-acetyltransferase activity / regulation of transcription elongation by RNA polymerase II / embryonic cleavage / box C/D methylation guide snoRNP complex / tRNA export from nucleus / rRNA base methylation / spindle assembly involved in female meiosis / cilium disassembly / epigenetic programming in the zygotic pronuclei / transcription elongation factor activity / rRNA primary transcript binding / Cul4-RING E3 ubiquitin ligase complex / blastocyst formation / sno(s)RNA-containing ribonucleoprotein complex / RNA splicing, via transesterification reactions / protein localization to nucleolus / U4 snRNA binding / negative regulation of RNA splicing / SUMOylation of RNA binding proteins / telomerase holoenzyme complex / rRNA methylation / U2-type precatalytic spliceosome / U3 snoRNA binding / box C/D snoRNP assembly / neural crest cell differentiation / rRNA modification in the nucleus and cytosol / erythrocyte homeostasis / Formation of the ternary complex, and subsequently, the 43S complex / cytoplasmic side of rough endoplasmic reticulum membrane / preribosome, small subunit precursor / negative regulation of ubiquitin protein ligase activity / Ribosomal scanning and start codon recognition / NRAGE signals death through JNK / snoRNA binding / precatalytic spliceosome / Translation initiation complex formation / intercellular bridge / postsynaptic modulation of chemical synaptic transmission / positive regulation of transcription by RNA polymerase I / mammalian oogenesis stage / protein acetylation / activation-induced cell death of T cells / Protein hydroxylation / RNA polymerase II complex binding / Association of TriC/CCT with target proteins during biosynthesis / mTORC1-mediated signalling / SARS-CoV-1 modulates host translation machinery / Peptide chain elongation / positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator / TFIID-class transcription factor complex binding / Selenocysteine synthesis / negative regulation of telomere maintenance via telomerase / Formation of a pool of free 40S subunits / blastocyst development / ubiquitin ligase inhibitor activity / Eukaryotic Translation Termination / negative regulation of apoptotic signaling pathway / decidualization / Response of EIF2AK4 (GCN2) to amino acid deficiency / SRP-dependent cotranslational protein targeting to membrane / Viral mRNA Translation / chromosome, centromeric region / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
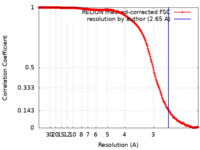
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.65 Å | |||||||||
 データ登録者 データ登録者 | Vanden Broeck A / Singh S / Klinge S | |||||||||
| 資金援助 |  米国, 2件 米国, 2件
| |||||||||
 引用 引用 |  ジャーナル: Science / 年: 2021 ジャーナル: Science / 年: 2021タイトル: Nucleolar maturation of the human small subunit processome. 著者: Sameer Singh / Arnaud Vanden Broeck / Linamarie Miller / Malik Chaker-Margot / Sebastian Klinge /  要旨: The human small subunit processome mediates early maturation of the small ribosomal subunit by coupling RNA folding to subsequent RNA cleavage and processing steps. We report the high-resolution ...The human small subunit processome mediates early maturation of the small ribosomal subunit by coupling RNA folding to subsequent RNA cleavage and processing steps. We report the high-resolution cryo–electron microscopy structures of maturing human small subunit (SSU) processomes at resolutions of 2.7 to 3.9 angstroms. These structures reveal the molecular mechanisms that enable crucial progressions during SSU processome maturation. RNA folding states within these particles are communicated to and coordinated with key enzymes that drive irreversible steps such as targeted exosome-mediated RNA degradation, protein-guided site-specific endonucleolytic RNA cleavage, and tightly controlled RNA unwinding. These conserved mechanisms highlight the SSU processome’s impressive structural plasticity, which endows this 4.5-megadalton nucleolar assembly with the distinctive ability to mature the small ribosomal subunit from within. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_23940.map.gz emd_23940.map.gz | 66.5 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-23940-v30.xml emd-23940-v30.xml emd-23940.xml emd-23940.xml | 18.1 KB 18.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_23940_fsc.xml emd_23940_fsc.xml | 19.7 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_23940.png emd_23940.png | 169.4 KB | ||
| マスクデータ |  emd_23940_msk_1.map emd_23940_msk_1.map | 669.9 MB |  マスクマップ マスクマップ | |
| その他 |  emd_23940_additional_1.map.gz emd_23940_additional_1.map.gz emd_23940_half_map_1.map.gz emd_23940_half_map_1.map.gz emd_23940_half_map_2.map.gz emd_23940_half_map_2.map.gz | 627 MB 542 MB 541.9 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23940 http://ftp.pdbj.org/pub/emdb/structures/EMD-23940 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23940 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23940 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_23940_validation.pdf.gz emd_23940_validation.pdf.gz | 457.5 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_23940_full_validation.pdf.gz emd_23940_full_validation.pdf.gz | 457.1 KB | 表示 | |
| XML形式データ |  emd_23940_validation.xml.gz emd_23940_validation.xml.gz | 27.4 KB | 表示 | |
| CIF形式データ |  emd_23940_validation.cif.gz emd_23940_validation.cif.gz | 36.6 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23940 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23940 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23940 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23940 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  7mq8C  7mq9C  7mqaC  7mqjC C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | |
| 電子顕微鏡画像生データ |  EMPIAR-10781 (タイトル: Nucleolar maturation of the human small subunit processome EMPIAR-10781 (タイトル: Nucleolar maturation of the human small subunit processomeData size: 74.6 TB Data #1: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 1 [micrographs - multiframe] Data #2: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 2 [micrographs - multiframe] Data #3: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 3 [micrographs - multiframe] Data #4: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 4 [micrographs - multiframe] Data #5: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 5 [micrographs - multiframe] Data #6: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 6 [micrographs - multiframe] Data #7: Aligned and averaged micrographs of human SSU processomes - Dataset 1 [micrographs - single frame] Data #8: Aligned and averaged micrographs of human SSU processomes - Dataset 2 [micrographs - single frame] Data #9: Aligned and averaged micrographs of human SSU processomes - Dataset 3 [micrographs - single frame] Data #10: Aligned and averaged micrographs of human SSU processomes - Dataset 4 [micrographs - single frame] Data #11: Aligned and averaged micrographs of human SSU processomes - Dataset 5 [micrographs - single frame] Data #12: Aligned and averaged micrographs of human SSU processomes - Dataset 6 [micrographs - single frame]) |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_23940.map.gz / 形式: CCP4 / 大きさ: 669.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_23940.map.gz / 形式: CCP4 / 大きさ: 669.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
-マスク #1
| ファイル |  emd_23940_msk_1.map emd_23940_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-追加マップ: #1
| ファイル | emd_23940_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Half-map1
| ファイル | emd_23940_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half-map1 | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Half-map2
| ファイル | emd_23940_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half-map2 | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Human SSU processome, state post-A1
| 全体 | 名称: Human SSU processome, state post-A1 |
|---|---|
| 要素 |
|
-超分子 #1: Human SSU processome, state post-A1
| 超分子 | 名称: Human SSU processome, state post-A1 / タイプ: complex / ID: 1 / 親要素: 0 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 5 MDa |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.6 |
|---|---|
| グリッド | モデル: Quantifoil R2/2 / 材質: GOLD / メッシュ: 400 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: HOLEY / 支持フィルム - Film thickness: 3.0 nm / 前処理 - タイプ: GLOW DISCHARGE |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 90 % / チャンバー内温度: 283 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 特殊光学系 | エネルギーフィルター - スリット幅: 20 eV |
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 実像数: 84904 / 平均電子線量: 58.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 0.01 mm / 最大 デフォーカス(公称値): 2.7 µm 最小 デフォーカス(公称値): 0.7000000000000001 µm |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 精密化 | 空間: REAL |
|---|
 ムービー
ムービー コントローラー
コントローラー









































































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)