+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7egd | ||||||
|---|---|---|---|---|---|---|---|
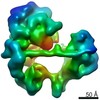
| Title | SCP promoter-bound TFIID-TFIIA in initial TBP-loading state | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSCRIPTION / TFIID / preinitiation complex / core promoter / transcription initiation | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of MHC class I biosynthetic process / spermine transport / SAGA complex assembly / lateral mesodermal cell differentiation / DNA-templated transcription open complex formation / allantois development / pre-snoRNP complex / positive regulation of androgen receptor signaling pathway / TFIIH-class transcription factor complex binding / negative regulation of protein autoubiquitination ...negative regulation of MHC class I biosynthetic process / spermine transport / SAGA complex assembly / lateral mesodermal cell differentiation / DNA-templated transcription open complex formation / allantois development / pre-snoRNP complex / positive regulation of androgen receptor signaling pathway / TFIIH-class transcription factor complex binding / negative regulation of protein autoubiquitination / negative regulation of MHC class II biosynthetic process / transcription factor TFTC complex / RNA polymerase I general transcription initiation factor activity / regulation of cell cycle G1/S phase transition / RNA polymerase transcription factor SL1 complex / histone H4K16ac reader activity / SLIK (SAGA-like) complex / RNA polymerase III general transcription initiation factor activity / RNA polymerase I core promoter sequence-specific DNA binding / hepatocyte differentiation / positive regulation of response to cytokine stimulus / RNA Polymerase III Transcription Initiation From Type 1 Promoter / RNA Polymerase III Transcription Initiation From Type 2 Promoter / RNA Polymerase III Transcription Initiation From Type 3 Promoter / transcription factor TFIIA complex / maintenance of protein location in nucleus / C2H2 zinc finger domain binding / female germ cell nucleus / RNA Polymerase III Abortive And Retractive Initiation / histone H3K4me3 reader activity / male pronucleus / host-mediated activation of viral transcription / female pronucleus / RNA polymerase II general transcription initiation factor binding / nuclear vitamin D receptor binding / RNA polymerase binding / regulation of fat cell differentiation / nuclear thyroid hormone receptor binding / limb development / box C/D snoRNP assembly / SAGA complex / transcription preinitiation complex / RNA Polymerase I Transcription Termination / inner cell mass cell proliferation / RNA polymerase II general transcription initiation factor activity / negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / transcription factor TFIID complex / histone acetyltransferase binding / HIV Transcription Initiation / RNA Polymerase II HIV Promoter Escape / Transcription of the HIV genome / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / midbrain development / cellular response to ATP / regulation of RNA splicing / negative regulation of signal transduction by p53 class mediator / negative regulation of cell cycle / aryl hydrocarbon receptor binding / transcription initiation at RNA polymerase I promoter / ubiquitin conjugating enzyme activity / TFIIB-class transcription factor binding / P-TEFb complex binding / RNA Polymerase I Transcription Initiation / MLL1 complex / transcription by RNA polymerase III / RNA polymerase II transcribes snRNA genes / negative regulation of ubiquitin-dependent protein catabolic process / positive regulation of transcription initiation by RNA polymerase II / embryonic placenta development / somitogenesis / histone acetyltransferase activity / core promoter sequence-specific DNA binding / RNA polymerase II core promoter sequence-specific DNA binding / regulation of DNA repair / negative regulation of protein kinase activity / RNA polymerase II preinitiation complex assembly / histone acetyltransferase / transcription regulator inhibitor activity / ovarian follicle development / estrogen receptor signaling pathway / positive regulation of intrinsic apoptotic signaling pathway / RNA Polymerase II Pre-transcription Events / negative regulation of proteasomal ubiquitin-dependent protein catabolic process / response to interleukin-1 / TBP-class protein binding / regulation of signal transduction by p53 class mediator / nuclear estrogen receptor binding / nuclear receptor binding / SIRT1 negatively regulates rRNA expression / male germ cell nucleus / transcription initiation at RNA polymerase II promoter / promoter-specific chromatin binding / RNA Polymerase I Promoter Escape / DNA-templated transcription initiation / mRNA transcription by RNA polymerase II / G1/S transition of mitotic cell cycle / euchromatin Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 6.75 Å | ||||||
 Authors Authors | Chen, X. / Wu, Z. / Li, J. / Zhao, D. / Xu, Y. | ||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Structural insights into preinitiation complex assembly on core promoters. Authors: Xizi Chen / Yilun Qi / Zihan Wu / Xinxin Wang / Jiabei Li / Dan Zhao / Haifeng Hou / Yan Li / Zishuo Yu / Weida Liu / Mo Wang / Yulei Ren / Ze Li / Huirong Yang / Yanhui Xu /  Abstract: Transcription factor IID (TFIID) recognizes core promoters and supports preinitiation complex (PIC) assembly for RNA polymerase II (Pol II)-mediated eukaryotic transcription. We determined the ...Transcription factor IID (TFIID) recognizes core promoters and supports preinitiation complex (PIC) assembly for RNA polymerase II (Pol II)-mediated eukaryotic transcription. We determined the structures of human TFIID-based PIC in three stepwise assembly states and revealed two-track PIC assembly: stepwise promoter deposition to Pol II and extensive modular reorganization on track I (on TATA-TFIID-binding element promoters) versus direct promoter deposition on track II (on TATA-only and TATA-less promoters). The two tracks converge at an ~50-subunit holo PIC in identical conformation, whereby TFIID stabilizes PIC organization and supports loading of cyclin-dependent kinase (CDK)-activating kinase (CAK) onto Pol II and CAK-mediated phosphorylation of the Pol II carboxyl-terminal domain. Unexpectedly, TBP of TFIID similarly bends TATA box and TATA-less promoters in PIC. Our study provides structural visualization of stepwise PIC assembly on highly diversified promoters. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7egd.cif.gz 7egd.cif.gz | 1.1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7egd.ent.gz pdb7egd.ent.gz | 868.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7egd.json.gz 7egd.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7egd_validation.pdf.gz 7egd_validation.pdf.gz | 927.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7egd_full_validation.pdf.gz 7egd_full_validation.pdf.gz | 942.3 KB | Display | |
| Data in XML |  7egd_validation.xml.gz 7egd_validation.xml.gz | 122.7 KB | Display | |
| Data in CIF |  7egd_validation.cif.gz 7egd_validation.cif.gz | 198 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/eg/7egd https://data.pdbj.org/pub/pdb/validation_reports/eg/7egd ftp://data.pdbj.org/pub/pdb/validation_reports/eg/7egd ftp://data.pdbj.org/pub/pdb/validation_reports/eg/7egd | HTTPS FTP |
-Related structure data
| Related structure data |  31113MC  7edxC  7eg7C  7eg8C  7eg9C  7egaC  7egbC  7egcC  7egeC  7egfC  7eggC  7eghC  7egiC  7egjC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Transcription initiation factor TFIID subunit ... , 13 types, 19 molecules ABDdEeFfGHIiJjLlckm
| #1: Protein | Mass: 212956.172 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TAF1, BA2R, CCG1, CCGS, TAF2A / Production host: Homo sapiens (human) / Gene: TAF1, BA2R, CCG1, CCGS, TAF2A / Production host:  Homo sapiens (human) Homo sapiens (human)References: UniProt: P21675, histone acetyltransferase, non-specific serine/threonine protein kinase | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #2: Protein | Mass: 137159.984 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TAF2, CIF150, TAF2B / Production host: Homo sapiens (human) / Gene: TAF2, CIF150, TAF2B / Production host:  Homo sapiens (human) / References: UniProt: Q6P1X5 Homo sapiens (human) / References: UniProt: Q6P1X5 | ||||||||||||||||||||
| #3: Protein | Mass: 110221.883 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 / Production host: Homo sapiens (human) / Gene: TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 / Production host:  Homo sapiens (human) / References: UniProt: O00268 Homo sapiens (human) / References: UniProt: O00268#4: Protein | Mass: 86932.109 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TAF5, TAF2D / Production host: Homo sapiens (human) / Gene: TAF5, TAF2D / Production host:  Homo sapiens (human) / References: UniProt: Q15542 Homo sapiens (human) / References: UniProt: Q15542#5: Protein | Mass: 72749.297 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TAF6, TAF2E, TAFII70 / Production host: Homo sapiens (human) / Gene: TAF6, TAF2E, TAFII70 / Production host:  Homo sapiens (human) / References: UniProt: P49848 Homo sapiens (human) / References: UniProt: P49848#6: Protein | | Mass: 40325.117 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TAF7, TAF2F, TAFII55 / Production host: Homo sapiens (human) / Gene: TAF7, TAF2F, TAFII55 / Production host:  Homo sapiens (human) / References: UniProt: Q15545 Homo sapiens (human) / References: UniProt: Q15545#7: Protein | | Mass: 34304.359 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TAF8, TAFII43, TBN / Production host: Homo sapiens (human) / Gene: TAF8, TAFII43, TBN / Production host:  Homo sapiens (human) / References: UniProt: Q7Z7C8 Homo sapiens (human) / References: UniProt: Q7Z7C8#8: Protein | Mass: 29006.838 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TAF9, TAF2G, TAFII31 / Production host: Homo sapiens (human) / Gene: TAF9, TAF2G, TAFII31 / Production host:  Homo sapiens (human) / References: UniProt: Q16594 Homo sapiens (human) / References: UniProt: Q16594#9: Protein | Mass: 21731.248 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TAF10, TAF2A, TAF2H, TAFII30 / Production host: Homo sapiens (human) / Gene: TAF10, TAF2A, TAF2H, TAFII30 / Production host:  Homo sapiens (human) / References: UniProt: Q12962 Homo sapiens (human) / References: UniProt: Q12962#10: Protein | Mass: 17948.467 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TAF12, TAF15, TAF2J, TAFII20 / Production host: Homo sapiens (human) / Gene: TAF12, TAF15, TAF2J, TAFII20 / Production host:  Homo sapiens (human) / References: UniProt: Q16514 Homo sapiens (human) / References: UniProt: Q16514#16: Protein | | Mass: 103769.320 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TAF3 / Production host: Homo sapiens (human) / Gene: TAF3 / Production host:  Homo sapiens (human) / References: UniProt: Q5VWG9 Homo sapiens (human) / References: UniProt: Q5VWG9#17: Protein | | Mass: 23340.094 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TAF11, TAF2I, PRO2134 / Production host: Homo sapiens (human) / Gene: TAF11, TAF2I, PRO2134 / Production host:  Homo sapiens (human) / References: UniProt: Q15544 Homo sapiens (human) / References: UniProt: Q15544#18: Protein | | Mass: 14307.068 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TAF13, TAF2K, TAFII18 / Production host: Homo sapiens (human) / Gene: TAF13, TAF2K, TAFII18 / Production host:  Homo sapiens (human) / References: UniProt: Q15543 Homo sapiens (human) / References: UniProt: Q15543 |
-Transcription initiation factor IIA subunit ... , 2 types, 2 molecules OQ
| #11: Protein | Mass: 12469.091 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GTF2A2, TF2A2 / Production host: Homo sapiens (human) / Gene: GTF2A2, TF2A2 / Production host:  Homo sapiens (human) / References: UniProt: P52657 Homo sapiens (human) / References: UniProt: P52657 |
|---|---|
| #13: Protein | Mass: 41544.551 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GTF2A1, TF2A1 / Production host: Homo sapiens (human) / Gene: GTF2A1, TF2A1 / Production host:  Homo sapiens (human) / References: UniProt: P52655 Homo sapiens (human) / References: UniProt: P52655 |
-Protein , 1 types, 1 molecules P
| #12: Protein | Mass: 37729.938 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TBP, GTF2D1, TF2D, TFIID / Production host: Homo sapiens (human) / Gene: TBP, GTF2D1, TF2D, TFIID / Production host:  Homo sapiens (human) / References: UniProt: P20226 Homo sapiens (human) / References: UniProt: P20226 |
|---|
-DNA chain , 2 types, 2 molecules XY
| #14: DNA chain | Mass: 22317.238 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Production host: Homo sapiens (human) / Production host:  Homo sapiens (human) Homo sapiens (human) |
|---|---|
| #15: DNA chain | Mass: 22090.072 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Production host: Homo sapiens (human) / Production host:  Homo sapiens (human) Homo sapiens (human) |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: SCP promoter-bound TFIID-TFIIA in initial TBP-loading state Type: COMPLEX / Entity ID: all / Source: RECOMBINANT |
|---|---|
| Molecular weight | Units: KILODALTONS/NANOMETER / Experimental value: NO |
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Buffer solution | pH: 7.9 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Grid material: GOLD / Grid type: Quantifoil R1.2/1.3 |
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 50 e/Å2 / Detector mode: SUPER-RESOLUTION / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
| Image scans | Movie frames/image: 32 |
- Processing
Processing
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
|---|---|
| 3D reconstruction | Resolution: 6.75 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 100170 / Symmetry type: POINT |
 Movie
Movie Controller
Controller

































 PDBj
PDBj








































