+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3744 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pronase-treated straight filament in Alzheimer's disease brain | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Human (human) Human (human) | |||||||||
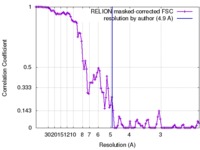
| Method | helical reconstruction / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Fitzpatrick AWP / Falcon B / He S / Murzin AG / Murshudov G / Garringer HG / Crowther RA / Ghetti B / Goedert M / Scheres SHW | |||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: Cryo-EM structures of tau filaments from Alzheimer's disease. Authors: Anthony W P Fitzpatrick / Benjamin Falcon / Shaoda He / Alexey G Murzin / Garib Murshudov / Holly J Garringer / R Anthony Crowther / Bernardino Ghetti / Michel Goedert / Sjors H W Scheres /   Abstract: Alzheimer's disease is the most common neurodegenerative disease, and there are no mechanism-based therapies. The disease is defined by the presence of abundant neurofibrillary lesions and neuritic ...Alzheimer's disease is the most common neurodegenerative disease, and there are no mechanism-based therapies. The disease is defined by the presence of abundant neurofibrillary lesions and neuritic plaques in the cerebral cortex. Neurofibrillary lesions comprise paired helical and straight tau filaments, whereas tau filaments with different morphologies characterize other neurodegenerative diseases. No high-resolution structures of tau filaments are available. Here we present cryo-electron microscopy (cryo-EM) maps at 3.4-3.5 Å resolution and corresponding atomic models of paired helical and straight filaments from the brain of an individual with Alzheimer's disease. Filament cores are made of two identical protofilaments comprising residues 306-378 of tau protein, which adopt a combined cross-β/β-helix structure and define the seed for tau aggregation. Paired helical and straight filaments differ in their inter-protofilament packing, showing that they are ultrastructural polymorphs. These findings demonstrate that cryo-EM allows atomic characterization of amyloid filaments from patient-derived material, and pave the way for investigation of a range of neurodegenerative diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3744.map.gz emd_3744.map.gz | 4.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3744-v30.xml emd-3744-v30.xml emd-3744.xml emd-3744.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3744_fsc.xml emd_3744_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_3744.png emd_3744.png | 35.9 KB | ||
| Others |  emd_3744_half_map_1.map.gz emd_3744_half_map_1.map.gz emd_3744_half_map_2.map.gz emd_3744_half_map_2.map.gz | 58.3 MB 66.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3744 http://ftp.pdbj.org/pub/emdb/structures/EMD-3744 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3744 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3744 | HTTPS FTP |
-Validation report
| Summary document |  emd_3744_validation.pdf.gz emd_3744_validation.pdf.gz | 352.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3744_full_validation.pdf.gz emd_3744_full_validation.pdf.gz | 351.5 KB | Display | |
| Data in XML |  emd_3744_validation.xml.gz emd_3744_validation.xml.gz | 15.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3744 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3744 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3744 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3744 | HTTPS FTP |
-Related structure data
| Related structure data |  3741C  3742C  3743C  5o3lC  5o3oC  5o3tC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3744.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3744.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_3744_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_3744_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tau
| Entire | Name: Tau |
|---|---|
| Components |
|
-Supramolecule #1: Tau
| Supramolecule | Name: Tau / type: tissue / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Tau
| Macromolecule | Name: Tau / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human (human) Human (human) |
| Sequence | String: VQIVYKPVDL SKVTSKCGSL GNIHHKPGGG QVEVKSEKLD FKDRVQSKIG SLDNITHVPG GGNKKIETHK LTF |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | tissue |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 20 mM Tris-HCl pH 7.4 containing 100 MM NaCl |
| Grid | Model: Quanitifoil Au R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
| Details | Sarkosyl-insoluble material was extracted from grey matter of frontal and temporal cortex from the patients brain and treated with pronase, as described in the Methods section of the paper. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum / Energy filter - Lower energy threshold: -10 eV / Energy filter - Upper energy threshold: 10 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 2-20 / Number grids imaged: 1 / Number real images: 523 / Average exposure time: 0.8 sec. / Average electron dose: 2.5 e/Å2 Details: images were collected in movie-mode at 1.2 frames per second |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 0.9 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.9 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 226-242 |
|---|---|
| Details | Fourier-space refinement of the complete atomic model against the paired helical filament and straight filament maps was performed in REFMAC. A stack of three consecutive monomers from each of the protofilaments was refined to preserve nearest-neighbour interactions for the middle chain. |
| Refinement | Space: RECIPROCAL / Protocol: AB INITIO MODEL / Overall B value: 79 / Target criteria: Fourier shell correlation |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)