-Search query
-Search result
Showing all 50 items for (author: wright & nj)

PDB-8yy8: 
Fzd7 -Gs complex

EMDB-41730: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Ribavirin
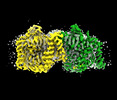
EMDB-41731: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in MSP2N2 nanodiscs, apo state
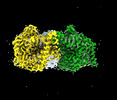
EMDB-41732: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with GS-441524, consensus reconstruction
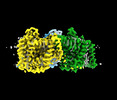
EMDB-41733: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with GS-441524, subset reconstruction
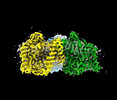
EMDB-41734: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with N-hydroxycytidine
EMDB-41735: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with PSI-6206

EMDB-41736: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT1-INT1-INT1 conformation
EMDB-41737: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT1-INT1-INT3 conformation
EMDB-41738: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 2, INT2-INT2-INT2 conformation

EMDB-41739: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 2, INT2-INT1-INT1 conformation

EMDB-41740: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, consensus reconstruction

EMDB-41741: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, OFS-OFS-OFS conformation

EMDB-41742: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, OFS-OFS-INT1 conformation

EMDB-41743: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, OFS-OFS-INT3 conformation
EMDB-41744: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT1-INT1-OFS conformation (from ensemble analysis)
EMDB-41745: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT3-INT3-OFS conformation
EMDB-41746: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT3-INT3-INT1 conformation
EMDB-41747: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT3-INT3-INT3 conformation
EMDB-41748: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, OFS-INT1-INT3 (clockwise) conformation
EMDB-41749: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, OFS-INT1-INT3 (counterclockwise) conformation

EMDB-41750: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 2, INT2-INT2-INT1 conformation

EMDB-41751: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 2, INT1-INT1-INT1 conformation

EMDB-41752: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 2, consensus reconstruction
EMDB-41755: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT1-INT1-OFS conformation (3DVA analysis)

EMDB-28586: 
Cryo-EM structure of the organic cation transporter 1 in the apo state

EMDB-28587: 
Cryo-EM structure of the organic cation transporter 1 in complex with diphenhydramine

EMDB-28588: 
Cryo-EM structure of the organic cation transporter 1 in complex with verapamil

EMDB-28589: 
Cryo-EM structure of the organic cation transporter 2 in complex with 1-methyl-4-phenylpyridinium

EMDB-26155: 
Cryo-EM structure of the human reduced folate carrier in complex with methotrexate

EMDB-26156: 
Cryo-EM structure of the human reduced folate carrier

EMDB-27394: 
Cryo-EM structure of the human reduced folate carrier, apo condition
EMDB-14132: 
Bovine complex I in lipid nanodisc, Active-Q10
EMDB-14133: 
Bovine complex I in lipid nanodisc, Active-apo
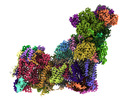
EMDB-14134: 
Bovine complex I in lipid nanodisc, Deactive-ligand (composite)
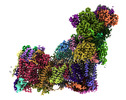
EMDB-14139: 
Bovine complex I in lipid nanodisc, Deactive-apo
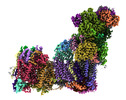
EMDB-14140: 
Bovine complex I in lipid nanodisc, State 3 (Slack)

PDB-7qsk: 
Bovine complex I in lipid nanodisc, Active-Q10

PDB-7qsl: 
Bovine complex I in lipid nanodisc, Active-apo

PDB-7qsm: 
Bovine complex I in lipid nanodisc, Deactive-ligand (composite)

PDB-7qsn: 
Bovine complex I in lipid nanodisc, Deactive-apo

PDB-7qso: 
Bovine complex I in lipid nanodisc, State 3 (Slack)

EMDB-23763: 
The F1 region of ammocidin-bound Saccharomyces cerevisiae ATP synthase

EMDB-23764: 
The F1 region of apoptolidin-bound Saccharomyces cerevisiae ATP synthase

EMDB-23765: 
The F1 region of inhibitor-free Saccharomyces cerevisiae ATP synthase

PDB-7md2: 
The F1 region of ammocidin-bound Saccharomyces cerevisiae ATP synthase

PDB-7md3: 
The F1 region of apoptolidin-bound Saccharomyces cerevisiae ATP synthase

EMDB-31340: 
Fzd7 -Gs complex

EMDB-12749: 
In situ cryo-electron tomogram of a pyrenoid inside a Chlamydomonas reinhardtii cell

EMDB-3694: 
In situ subtomogram average of Rubisco within the Chlamydomonas pyrenoid
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
