[English] 日本語
 Yorodumi
Yorodumi- EMDB-3694: In situ subtomogram average of Rubisco within the Chlamydomonas p... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3694 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ subtomogram average of Rubisco within the Chlamydomonas pyrenoid | |||||||||
 Map data Map data | In situ subtomogram average of Rubisco holoenzymes within the pyrenoid of Chlamydomonas reinhardtii. Filtered to 16%u212B. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 16.5 Å | |||||||||
 Authors Authors | Cuellar LK / Schaffer M / Strauss M / Martinez-Sanchez A / Plitzko JM / Foerster F / Engel BD | |||||||||
 Citation Citation |  Journal: Cell / Year: 2017 Journal: Cell / Year: 2017Title: The Eukaryotic CO-Concentrating Organelle Is Liquid-like and Exhibits Dynamic Reorganization. Authors: Elizabeth S Freeman Rosenzweig / Bin Xu / Luis Kuhn Cuellar / Antonio Martinez-Sanchez / Miroslava Schaffer / Mike Strauss / Heather N Cartwright / Pierre Ronceray / Jürgen M Plitzko / ...Authors: Elizabeth S Freeman Rosenzweig / Bin Xu / Luis Kuhn Cuellar / Antonio Martinez-Sanchez / Miroslava Schaffer / Mike Strauss / Heather N Cartwright / Pierre Ronceray / Jürgen M Plitzko / Friedrich Förster / Ned S Wingreen / Benjamin D Engel / Luke C M Mackinder / Martin C Jonikas /   Abstract: Approximately 30%-40% of global CO fixation occurs inside a non-membrane-bound organelle called the pyrenoid, which is found within the chloroplasts of most eukaryotic algae. The pyrenoid matrix is ...Approximately 30%-40% of global CO fixation occurs inside a non-membrane-bound organelle called the pyrenoid, which is found within the chloroplasts of most eukaryotic algae. The pyrenoid matrix is densely packed with the CO-fixing enzyme Rubisco and is thought to be a crystalline or amorphous solid. Here, we show that the pyrenoid matrix of the unicellular alga Chlamydomonas reinhardtii is not crystalline but behaves as a liquid that dissolves and condenses during cell division. Furthermore, we show that new pyrenoids are formed both by fission and de novo assembly. Our modeling predicts the existence of a "magic number" effect associated with special, highly stable heterocomplexes that influences phase separation in liquid-like organelles. This view of the pyrenoid matrix as a phase-separated compartment provides a paradigm for understanding its structure, biogenesis, and regulation. More broadly, our findings expand our understanding of the principles that govern the architecture and inheritance of liquid-like organelles. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3694.map.gz emd_3694.map.gz | 638.3 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3694-v30.xml emd-3694-v30.xml emd-3694.xml emd-3694.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
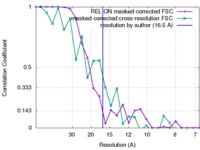
| FSC (resolution estimation) |  emd_3694_fsc_1.xml emd_3694_fsc_1.xml emd_3694_fsc_2.xml emd_3694_fsc_2.xml | 2.1 KB 2.4 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_3694.png emd_3694.png | 66.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3694 http://ftp.pdbj.org/pub/emdb/structures/EMD-3694 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3694 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3694 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3694.map.gz / Format: CCP4 / Size: 686.5 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3694.map.gz / Format: CCP4 / Size: 686.5 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | In situ subtomogram average of Rubisco holoenzymes within the pyrenoid of Chlamydomonas reinhardtii. Filtered to 16%u212B. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.42 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : In situ Rubisco holoenzyme
| Entire | Name: In situ Rubisco holoenzyme |
|---|---|
| Components |
|
-Supramolecule #1: In situ Rubisco holoenzyme
| Supramolecule | Name: In situ Rubisco holoenzyme / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: In situ subtomogram average generated from Rubisco holoenzymes imaged within the native Chlamydomonas pyrenoid. Cells were thinned by focused ion beam milling. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 540 KDa |
-Macromolecule #1: Rubisco holoenzyme
| Macromolecule | Name: Rubisco holoenzyme / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: ribulose-bisphosphate carboxylase |
|---|---|
| Sequence | String: MVPQTETKAG AGFKAGVKDY RLTYYTPDYV VRDTDILAAF RMTPQPGVPP EECGAAVAAE SSTGTWTTVW TDGLTSLDRY KGRCYDIEPV PGEDNQYIAY VAYPIDLFEE GSVTNMFTSI VGNVFGFKAL RALRLEDLRI PPAYVKTFVG PPHGIQVERD KLNKYGRGLL ...String: MVPQTETKAG AGFKAGVKDY RLTYYTPDYV VRDTDILAAF RMTPQPGVPP EECGAAVAAE SSTGTWTTVW TDGLTSLDRY KGRCYDIEPV PGEDNQYIAY VAYPIDLFEE GSVTNMFTSI VGNVFGFKAL RALRLEDLRI PPAYVKTFVG PPHGIQVERD KLNKYGRGLL GCTIKPKLGL SAKNYGRAVY ECLRGGLDFT KDDENVNSQP FMRWRDRFLF VAEAIYKAQA ETGEVKGHYL NATAGTCEEM MKRAVCAKEL GVPIIMHDYL TGGFTANTSL AIYCRDNGLL LHIHRAMHAV IDRQRNHGIH FRVLAKALRM SGGDHLHSGT VVGKLEGERE VTLGFVDLMR DDYVEKDRSR GIYFTQDWCS MPGVMPVASG GIHVWHMPAL VEIFGDDACL QFGGGTLGHP WGNAPGAAAN RVALEACTQA RNEGRDLARE GGDVIRSACK WSPELAAACE VWKEIKFEFD TIDKL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV Details: Blotted for 10 seconds with 10 blot force before plunging.. |
| Details | Rubisco holoenzymes within the native Chlamydomonas pyrenoid. Whole cells were plunge-frozen onto EM grids and then thinned with a focused ion beam instrument. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 1 / Average exposure time: 1.5 sec. / Average electron dose: 1.5 e/Å2 Details: Images were collected in movie mode at 17 frames per second |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 6.457 µm / Calibrated defocus min: 5.077 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)