[English] 日本語
 Yorodumi
Yorodumi- EMDB-12749: In situ cryo-electron tomogram of a pyrenoid inside a Chlamydomon... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12749 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ cryo-electron tomogram of a pyrenoid inside a Chlamydomonas reinhardtii cell | |||||||||
 Map data Map data | In situ cryo-electron tomogram of a pyrenoid inside a Chlamydomonas reinhardtii cell (bin4). A denoised version of this tomogram is included in the bundle. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Cuellar LK / Schaffer M / Strauss M / Martinez-Sanchez A / Plitzko JM / Foerster F / Engel BD | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2017 Journal: Cell / Year: 2017Title: The Eukaryotic CO-Concentrating Organelle Is Liquid-like and Exhibits Dynamic Reorganization. Authors: Elizabeth S Freeman Rosenzweig / Bin Xu / Luis Kuhn Cuellar / Antonio Martinez-Sanchez / Miroslava Schaffer / Mike Strauss / Heather N Cartwright / Pierre Ronceray / Jürgen M Plitzko / ...Authors: Elizabeth S Freeman Rosenzweig / Bin Xu / Luis Kuhn Cuellar / Antonio Martinez-Sanchez / Miroslava Schaffer / Mike Strauss / Heather N Cartwright / Pierre Ronceray / Jürgen M Plitzko / Friedrich Förster / Ned S Wingreen / Benjamin D Engel / Luke C M Mackinder / Martin C Jonikas /   Abstract: Approximately 30%-40% of global CO fixation occurs inside a non-membrane-bound organelle called the pyrenoid, which is found within the chloroplasts of most eukaryotic algae. The pyrenoid matrix is ...Approximately 30%-40% of global CO fixation occurs inside a non-membrane-bound organelle called the pyrenoid, which is found within the chloroplasts of most eukaryotic algae. The pyrenoid matrix is densely packed with the CO-fixing enzyme Rubisco and is thought to be a crystalline or amorphous solid. Here, we show that the pyrenoid matrix of the unicellular alga Chlamydomonas reinhardtii is not crystalline but behaves as a liquid that dissolves and condenses during cell division. Furthermore, we show that new pyrenoids are formed both by fission and de novo assembly. Our modeling predicts the existence of a "magic number" effect associated with special, highly stable heterocomplexes that influences phase separation in liquid-like organelles. This view of the pyrenoid matrix as a phase-separated compartment provides a paradigm for understanding its structure, biogenesis, and regulation. More broadly, our findings expand our understanding of the principles that govern the architecture and inheritance of liquid-like organelles. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12749.map.gz emd_12749.map.gz | 329.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12749-v30.xml emd-12749-v30.xml emd-12749.xml emd-12749.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_12749.png emd_12749.png | 155.9 KB | ||
| Others |  emd_12749_additional_1.map.gz emd_12749_additional_1.map.gz | 319.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12749 http://ftp.pdbj.org/pub/emdb/structures/EMD-12749 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12749 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12749 | HTTPS FTP |
-Related structure data
| Related structure data |  3694C C: citing same article ( |
|---|---|
| EM raw data |  EMPIAR-10694 (Title: In situ cryo-electron tomogram of a pyrenoid inside a Chlamydomonas reinhardtii cell (tilt series) EMPIAR-10694 (Title: In situ cryo-electron tomogram of a pyrenoid inside a Chlamydomonas reinhardtii cell (tilt series)Data size: 5.8 Data #1: Phase-flipped and aligned tilt series of a pyrenoid inside a Chlamydomonas reinhardtii cell [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12749.map.gz / Format: CCP4 / Size: 762.2 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) Download / File: emd_12749.map.gz / Format: CCP4 / Size: 762.2 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | In situ cryo-electron tomogram of a pyrenoid inside a Chlamydomonas reinhardtii cell (bin4). A denoised version of this tomogram is included in the bundle. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 13.68 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Denoised tomogram by deconvolving with an ad-hoc Wiener filter.
| File | emd_12749_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Denoised tomogram by deconvolving with an ad-hoc Wiener filter. | ||||||||||||
| Projections & Slices |
| ||||||||||||
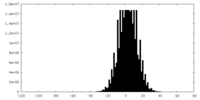
| Density Histograms |
- Sample components
Sample components
-Entire : Pyrenoid
| Entire | Name: Pyrenoid |
|---|---|
| Components |
|
-Supramolecule #1: Pyrenoid
| Supramolecule | Name: Pyrenoid / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: FEI VITROBOT MARK IV |
| Sectioning | Focused ion beam - Instrument: OTHER / Focused ion beam - Ion: OTHER / Focused ion beam - Voltage: 30 kV / Focused ion beam - Current: 0.03 nA / Focused ion beam - Duration: 1800 sec. / Focused ion beam - Temperature: 91 K / Focused ion beam - Initial thickness: 8000 nm / Focused ion beam - Final thickness: 300 nm Focused ion beam - Details: See https://bio-protocol.org/e1575 for detailed procedure.. The value given for _emd_sectioning_focused_ion_beam.instrument is FEI Quanta FIB. This is not in a list of ...Focused ion beam - Details: See https://bio-protocol.org/e1575 for detailed procedure.. The value given for _emd_sectioning_focused_ion_beam.instrument is FEI Quanta FIB. This is not in a list of allowed values {'OTHER', 'DB235'} so OTHER is written into the XML file. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Average exposure time: 1.0 sec. / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 42000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Software - Name:  IMOD / Number images used: 65 IMOD / Number images used: 65 |
|---|
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)