+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3853 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human apoferritin H | |||||||||
 Map data Map data | human apoferritin H | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationiron ion sequestering activity / ferritin complex / Scavenging by Class A Receptors / negative regulation of ferroptosis / Golgi Associated Vesicle Biogenesis / ferroxidase / autolysosome / ferroxidase activity / negative regulation of fibroblast proliferation / ferric iron binding ...iron ion sequestering activity / ferritin complex / Scavenging by Class A Receptors / negative regulation of ferroptosis / Golgi Associated Vesicle Biogenesis / ferroxidase / autolysosome / ferroxidase activity / negative regulation of fibroblast proliferation / ferric iron binding / autophagosome / iron ion transport / ferrous iron binding / Iron uptake and transport / tertiary granule lumen / ficolin-1-rich granule lumen / intracellular iron ion homeostasis / immune response / iron ion binding / negative regulation of cell population proliferation / Neutrophil degranulation / extracellular exosome / extracellular region / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Dudkina NV / Welsch S | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Falcon 3 Electron Counting detector enables high resolution cryo-EM structures Authors: Dudkina NV / Welsch S | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3853.map.gz emd_3853.map.gz | 9.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3853-v30.xml emd-3853-v30.xml emd-3853.xml emd-3853.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
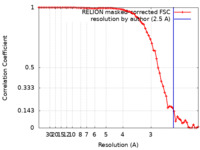
| FSC (resolution estimation) |  emd_3853_fsc.xml emd_3853_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_3853.png emd_3853.png | 64.3 KB | ||
| Others |  emd_3853_additional.map.gz emd_3853_additional.map.gz emd_3853_additional_1.map.gz emd_3853_additional_1.map.gz | 20.2 MB 20.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3853 http://ftp.pdbj.org/pub/emdb/structures/EMD-3853 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3853 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3853 | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3853.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3853.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human apoferritin H | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.057 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: #1
| File | emd_3853_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_3853_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human apoferritin H
| Entire | Name: Human apoferritin H |
|---|---|
| Components |
|
-Supramolecule #1: Human apoferritin H
| Supramolecule | Name: Human apoferritin H / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 509 KDa |
-Macromolecule #1: Ferritin heavy chain
| Macromolecule | Name: Ferritin heavy chain / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MTTASTSQVR QNYHQDSEA A INRQINLE LY ASYVYLS MSY YFDRDD VALK NFAKY FLHQS HEER EHAEKL MKL QNQRGGR IF LQDIKKPD C DDWESGLNA MECALHLEKN VNQSLLELH K LATDKNDP HL CDFIETH YLN EQVKAI KELGDHVTNL RKMG ...String: MTTASTSQVR QNYHQDSEA A INRQINLE LY ASYVYLS MSY YFDRDD VALK NFAKY FLHQS HEER EHAEKL MKL QNQRGGR IF LQDIKKPD C DDWESGLNA MECALHLEKN VNQSLLELH K LATDKNDP HL CDFIETH YLN EQVKAI KELGDHVTNL RKMG APESG LAEYL FDKH TLGDSD NES |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Component - Formula: PBS |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Protein was applied to glow-discharged Quantifoil R2/2 grids. |
| Details | The protein was provided by Institute of Biophysics of Chinese Academy of Sciences |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Alignment procedure | Coma free - Residual tilt: 10.0 mrad |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 14.0 µm / Number grids imaged: 1 / Number real images: 203 / Average exposure time: 55.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)