-Search query
-Search result
Showing all 19 items for (author: svergun & di)

EMDB-50408: 
Cryo-EM structure of Legionella effector SdeC (PDE-mART domain)
Method: single particle / : Weng TH, Misra M, Chen W, Safarian S, Kudryashev M, Dikic I

EMDB-50413: 
Cryo-EM structure of Legionella effector SdeC (3D flexible refinement)
Method: single particle / : Weng TH, Misra M, Chen W, Safarian S, Kudryashev M, Dikic I

EMDB-14618: 
Dipeptide and tripeptide Permease C (DtpC)
Method: single particle / : Killer M, Finocchio G, Pardon E, Steyaert J, Loew C

EMDB-11715: 
Cryo-EM reconstruction of the di-hexameric endocytic adaptor complex AENTH (ENTH F5A/L12A/V13A mutant)
Method: single particle / : Klebl DP, Lizarrondo J, Sobott F, Garcia-Alai MM, Muench SP

EMDB-11987: 
Structure of the endocytic adaptor complex AENTH
Method: single particle / : Klebl DP, Lizarrondo J

EMDB-11831: 
BIP18SN. De novo coiled-coil-based bipyramidal protein
Method: single particle / : Lapenta F, Aupic J, Vezzoli M, Strmsek Z, Da Vela S, Svergun DI, Melero R, Carazo JM, Jerala R

EMDB-11616: 
Cryo-EM structure of the SARS-CoV-2 spike protein bound to neutralizing sybodies (Sb23)
Method: single particle / : Hallberg BM, Das H

PDB-7a25: 
Cryo-EM structure of the SARS-CoV-2 spike protein bound to neutralizing sybodies (Sb23)
Method: single particle / : Hallberg BM, Das H

EMDB-11617: 
Cryo-EM structure of the SARS-CoV-2 spike protein bound to neutralizing sybodies (Sb23) 2-up conformation
Method: single particle / : Hallberg BM, Das H

PDB-7a29: 
Cryo-EM structure of the SARS-CoV-2 spike protein bound to neutralizing sybodies (Sb23) 2-up conformation
Method: single particle / : Hallberg BM, Das H

EMDB-3596: 
The structure of the ESX-5 mycobacterial type VII secretion system membrane complex
Method: single particle / : Ciccarelli L, Marlovits TC

EMDB-3502: 
Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes
Method: single particle / : Sundaramoorthy R, Owen-Hughes T

EMDB-3517: 
Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes.
Method: single particle / : Sundaramoorthy R, Owen-Hughes T
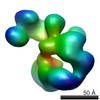
EMDB-2274: 
human crm1 extended conformation
Method: single particle / : Doelker N, Blanchet CE, Haselbach D, Voss B, Kappel C, Monecke T, Svergun D, Stark H, Ficner R, Zacharias U, Grubmueller H, Dickmanns A

EMDB-5564: 
Compact conformation of human crm1
Method: single particle / : Doelker N, Blanchet CE, Haselbach D, Voss B, Kappel C, Monecke T, Svergun D, Stark H, Ficner R, Zacharie U, Grubmueller H, Dickmanns A

EMDB-1977: 
Extracellular complexes of the hematopoietic human and mouse CSF-1 receptor are driven by common assembly principles.
Method: single particle / : Elegheert J, Desfosses A, Shkumatov AV, Wu X, Bracke N, Verstraete K, Van Craenenbroeck K, Brooks BR, Svergun DI, Vergauwen B, Gutsche I, Savvides SN

EMDB-1640: 
Structure of the V-ATPase of Saccharomyces cerevisiae at 2.5 nm resolution
Method: single particle / : Diepholz M, Venzke D, Prinz S, Batisse C, Florchinger B, Rossle M, Svergun D, Bottcher B, Fethiere J

EMDB-1440: 
The subnanometer resolution structure of the glutamate synthase 1.2-MDa hexamer by cryoelectron microscopy and its oligomerization behavior in solution: functional implications.
Method: single particle / : Cottevieille M, Larquet E, Jonic S, Petoukhov MV, Caprini G, Paravisi S, Svergun DI, Vanoni MA, Boisset N

EMDB-1003: 
Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Method: single particle / : Gabashvili IS
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
