[English] 日本語
 Yorodumi
Yorodumi- EMDB-1977: Extracellular complexes of the hematopoietic human and mouse CSF-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1977 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Extracellular complexes of the hematopoietic human and mouse CSF-1 receptor are driven by common assembly principles. | |||||||||
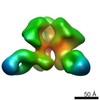
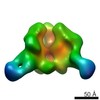
 Map data Map data | Surface rendering of the hCSF-1RD1-D5 hCSF-1 complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Hematopoiesis / Receptor Tyrosine Kinase (RTK) / Colony-Stimulating Factor-1 / CSF-1 / CSF-1R / ternary complex / ectodomain complex / cytokine-receptor complex | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Elegheert J / Desfosses A / Shkumatov AV / Wu X / Bracke N / Verstraete K / Van Craenenbroeck K / Brooks BR / Svergun DI / Vergauwen B ...Elegheert J / Desfosses A / Shkumatov AV / Wu X / Bracke N / Verstraete K / Van Craenenbroeck K / Brooks BR / Svergun DI / Vergauwen B / Gutsche I / Savvides SN | |||||||||
 Citation Citation |  Journal: Structure / Year: 2011 Journal: Structure / Year: 2011Title: Extracellular complexes of the hematopoietic human and mouse CSF-1 receptor are driven by common assembly principles. Authors: Jonathan Elegheert / Ambroise Desfosses / Alexander V Shkumatov / Xiongwu Wu / Nathalie Bracke / Kenneth Verstraete / Kathleen Van Craenenbroeck / Bernard R Brooks / Dmitri I Svergun / Bjorn ...Authors: Jonathan Elegheert / Ambroise Desfosses / Alexander V Shkumatov / Xiongwu Wu / Nathalie Bracke / Kenneth Verstraete / Kathleen Van Craenenbroeck / Bernard R Brooks / Dmitri I Svergun / Bjorn Vergauwen / Irina Gutsche / Savvas N Savvides /  Abstract: The hematopoietic colony stimulating factor-1 receptor (CSF-1R or FMS) is essential for the cellular repertoire of the mammalian immune system. Here, we report a structural and mechanistic consensus ...The hematopoietic colony stimulating factor-1 receptor (CSF-1R or FMS) is essential for the cellular repertoire of the mammalian immune system. Here, we report a structural and mechanistic consensus for the assembly of human and mouse CSF-1:CSF-1R complexes. The EM structure of the complete extracellular assembly of the human CSF-1:CSF-1R complex reveals how receptor dimerization by CSF-1 invokes a ternary complex featuring extensive homotypic receptor contacts and striking structural plasticity at the extremities of the complex. Studies by small-angle X-ray scattering of unliganded hCSF-1R point to large domain rearrangements upon CSF-1 binding, and provide structural evidence for the relevance of receptor predimerization at the cell surface. Comparative structural and binding studies aiming to dissect the assembly principles of human and mouse CSF-1R complexes, including a quantification of the CSF-1/CSF-1R species cross-reactivity, show that bivalent cytokine binding to receptor coupled to ensuing receptor-receptor interactions are common denominators in extracellular complex formation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1977.map.gz emd_1977.map.gz | 1.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1977-v30.xml emd-1977-v30.xml emd-1977.xml emd-1977.xml | 10.1 KB 10.1 KB | Display Display |  EMDB header EMDB header |
| Images |  1977_CSF1R_imagedeposition.png 1977_CSF1R_imagedeposition.png | 30 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1977 http://ftp.pdbj.org/pub/emdb/structures/EMD-1977 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1977 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1977 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1977.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1977.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Surface rendering of the hCSF-1RD1-D5 hCSF-1 complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of human Colony-Stimulating Factor-1 (hCSF-1) with the co...
| Entire | Name: Complex of human Colony-Stimulating Factor-1 (hCSF-1) with the complete ectodomain of hCSF-1R |
|---|---|
| Components |
|
-Supramolecule #1000: Complex of human Colony-Stimulating Factor-1 (hCSF-1) with the co...
| Supramolecule | Name: Complex of human Colony-Stimulating Factor-1 (hCSF-1) with the complete ectodomain of hCSF-1R type: sample / ID: 1000 / Details: The sample was monodisperse / Oligomeric state: Dimeric / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 145 KDa / Theoretical: 145 KDa / Method: Multi-angle laser light scattering |
-Macromolecule #1: Colony Stimulating Factor-1 Receptor (CSF-1R)
| Macromolecule | Name: Colony Stimulating Factor-1 Receptor (CSF-1R) / type: protein_or_peptide / ID: 1 / Name.synonym: CSF-1R / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Experimental: 76 KDa / Theoretical: 76 KDa |
| Recombinant expression | Organism: Homo sapiens, HEK293T cell line / Recombinant plasmid: pHLSec |
-Macromolecule #2: Colony Stimulating Factor-1
| Macromolecule | Name: Colony Stimulating Factor-1 / type: protein_or_peptide / ID: 2 / Name.synonym: CSF-1 / Number of copies: 2 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism: Homo sapiens, HEK293T cell line / Recombinant plasmid: pHLSec |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 20 mM NaPO4 pH 7.40, 150 mM NaCl. |
|---|---|
| Staining | Type: NEGATIVE Details: Purified sample at 0.2 mg/mL in PBS buffer was applied to the clear side of carbon on a carbon-mica interface and stained by floating on 2 % (w/v) uranyl acetate. |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1200EXII |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 14 µm / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 100 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.1 mm / Nominal magnification: 40000 |
| Sample stage | Specimen holder: Jeol / Specimen holder model: JEOL |
- Image processing
Image processing
| CTF correction | Details: CTFFIND3. Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: IMAGIC, SPIDER / Number images used: 9421 |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















