+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDAR6 |
|---|---|
 Sample Sample | human CSF-1:CSF-1R extracellular signalling complex
|
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Journal: Structure / Year: 2015 Journal: Structure / Year: 2015Title: Structure and Assembly Mechanism of the Signaling Complex Mediated by Human CSF-1. Authors: Jan Felix / Steven De Munck / Kenneth Verstraete / Leander Meuris / Nico Callewaert / Jonathan Elegheert / Savvas N Savvides /  Abstract: Human colony-stimulating factor 1 receptor (hCSF-1R) is unique among the hematopoietic receptors because it is activated by two distinct cytokines, CSF-1 and interleukin-34 (IL-34). Despite ever- ...Human colony-stimulating factor 1 receptor (hCSF-1R) is unique among the hematopoietic receptors because it is activated by two distinct cytokines, CSF-1 and interleukin-34 (IL-34). Despite ever-growing insights into the central role of hCSF-1R signaling in innate and adaptive immunity, inflammatory diseases, and cancer, the structural basis of the functional dichotomy of hCSF-1R has remained elusive. Here, we report crystal structures of ternary complexes between hCSF-1 and hCSF-1R, including their complete extracellular assembly, and propose a mechanism for the cooperative human CSF-1:CSF-1R complex that relies on the adoption by dimeric hCSF-1 of an active conformational state and homotypic receptor interactions. Furthermore, we trace the cytokine-binding duality of hCSF-1R to a limited set of conserved interactions mediated by functionally equivalent residues on CSF-1 and IL-34 that play into the geometric requirements of hCSF-1R activation, and map the possible mechanistic consequences of somatic mutations in hCSF-1R associated with cancer. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
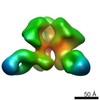
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Models
| Model #216 |  Type: mix / Software: SASREF / Symmetry: P2 / Chi-square value: 2.23603086735  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|
- Sample
Sample
 Sample Sample | Name: human CSF-1:CSF-1R extracellular signalling complex / Specimen concentration: 0.50-10.00 / Entity id: 130 / 131 |
|---|---|
| Buffer | Name: 50 mM NaH2PO4, 100 m / pH: 7.4 |
| Entity #130 | Name: hCSF-1 / Type: protein / Description: Macrophage colony-stimulating factor 1 / Formula weight: 17.436 / Num. of mol.: 2 / Source: Homo sapiens Sequence: EEVSEYCSHM IGSGHLQSLQ RLIDSQMETS CQITFEFVDQ EQLKDPVCYL KKAFLLVQDI MEDTMRFRDN TPNAIAIVQL QELSLRLKSC FTKDYEEHDK ACVRTFYETP LQLLEKVKNV FNETKNLLDK DWNIFSKNCN NSFAECSSQ |
| Entity #131 | Name: hCSF-1R / Type: protein Description: Macrophage colony-stimulating factor 1 receptor Formula weight: 53.554 / Num. of mol.: 2 / Source: Homo sapiens Sequence: IPVIEPSVPE LVVKPGATVT LRCVGNGSVE WDGPPSPHWT LYSDGSSSIL STNNATFQNT GTYRCTEPGD PLGGSAAIHL YVKDPARPWN VLAQEVVVFE DQDALLPCLL TDPVLEAGVS LVRVRGRPLM RHTNYSFSPW HGFTIHRAKF IQSQDYQCSA LMGGRKVMSI ...Sequence: IPVIEPSVPE LVVKPGATVT LRCVGNGSVE WDGPPSPHWT LYSDGSSSIL STNNATFQNT GTYRCTEPGD PLGGSAAIHL YVKDPARPWN VLAQEVVVFE DQDALLPCLL TDPVLEAGVS LVRVRGRPLM RHTNYSFSPW HGFTIHRAKF IQSQDYQCSA LMGGRKVMSI SIRLKVQKVI PGPPALTLVP AELVRIRGEA AQIVCSASSV DVNFDVFLQH NNTKLAIPQQ SDFHNNRYQK VLTLNLDQVD FQHAGNYSCV ASNVQGKHST SMFFRVVESA YLNLSSEQNL IQEVTVGEGL NLKVMVEAYP GLQGFNWTYL GPFSDHQPEP KLANATTKDT YRHTFTLSLP RLKPSEAGRY SFLARNPGGW RALTFELTLR YPPEVSVIWT FINGSGTLLC AASGYPQPNV TWLQCSGHTD RCDEAQVLQV WDDPYPEVLS QEPFHKVTVQ SLLTVETLEH NQTYECRAHN SVGSGSWAFI PISAGTKHHH HHH |
-Experimental information
| Beam | Instrument name:  DORIS III X33 DORIS III X33  / City: Hamburg / 国: Germany / City: Hamburg / 国: Germany  / Shape: 0.6 / Type of source: X-ray synchrotron / Wavelength: 0.15 Å / Dist. spec. to detc.: 2.7 mm / Shape: 0.6 / Type of source: X-ray synchrotron / Wavelength: 0.15 Å / Dist. spec. to detc.: 2.7 mm | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M-W / Pixsize x: 0.172 mm | |||||||||||||||||||||
| Scan |
| |||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||
| Result | Comments: SAXS data acquisition for the glycosylated ternary hCSF-1:hCSF-1R full ectodomain complex (MW including glycans: 158 kDa) and subsequent data processing were performed as described in ...Comments: SAXS data acquisition for the glycosylated ternary hCSF-1:hCSF-1R full ectodomain complex (MW including glycans: 158 kDa) and subsequent data processing were performed as described in Elegheert et al., 2011 and Felix et al., 2013 respectively (see references). Rigid-body refinements of the hCSF-1:hCSF-1R complex were performed using the online version of SASREF. For each run, data to 0.25 angstrom-1 was used while imposing P2 symmetry. The crystal structure of full length hCSF-1:hCSF-1R without D1 was taken as a starting rigid-body core (PDB ID: 4WRM). D1 was added as a separate rigid body with a contact restraint of 4 angstrom between its C-terminus and the N-terminus of hCSF-1RD2-D5, as well as 5 N-linked glycans containing a 'dummy' Asparagine residue (Asn-GlcNac2Man5) with 1 angstrom restraints to the C-alpha of truncated Asparagine residues on hCSF-1RD2-D5 (N73, N153, N240, N275 and N353). The SASREF calculated fit of the model to the experimental data gave a chi-squared of 1.65, recalculation of the fit using Crysol and FoXS gave a chi-squared of 3.0 and 1.5 respectively. Here, the fit calculated with FoXS is presented. Felix, J., Elegheert, J., Gutsche, I., Shkumatov, A.V., Wen, Y., Bracke, N., Pannecoucke, E., Vandenberghe, I., Devreese, B., Svergun, D.I., et al. (2013). Human IL-34 and CSF-1 establish structurally similar extracellular assemblies with their common hematopoietic receptor. Structure 21, 528-539. Elegheert, J., Desfosses, A., Shkumatov, A.V., Wu, X., Bracke, N., Verstraete, K., Van Craenenbroeck, K., Brooks, B.R., Svergun, D.I., Vergauwen, B., et al. (2011). Extracellular Complexes of the Hematopoietic Human and Mouse CSF-1 Receptor Are Driven by Common Assembly Principles. Structure 19, 1762-1772.
|
 Movie
Movie Controller
Controller


 SASDAR6
SASDAR6