+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Dipeptide and tripeptide Permease C (DtpC) | |||||||||
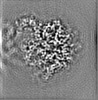
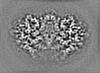
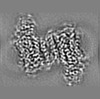
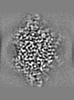
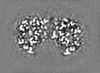
 Map data Map data | Post-processed map. Focused ref on one copy of DtpC. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane protein / Peptide transporter / Proton coupled oligopeptide transporter / POT / MFS / DtpC. / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationdipeptide transmembrane transporter activity / peptide transmembrane transporter activity / protein transport / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.72 Å | |||||||||
 Authors Authors | Killer M / Finocchio G / Pardon E / Steyaert J / Loew C | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Front Mol Biosci / Year: 2022 Journal: Front Mol Biosci / Year: 2022Title: Cryo-EM Structure of an Atypical Proton-Coupled Peptide Transporter: Di- and Tripeptide Permease C. Authors: Maxime Killer / Giada Finocchio / Haydyn D T Mertens / Dmitri I Svergun / Els Pardon / Jan Steyaert / Christian Löw /   Abstract: Proton-coupled Oligopeptide Transporters (POTs) of the Major Facilitator Superfamily (MFS) mediate the uptake of short di- and tripeptides in all phyla of life. POTs are thought to constitute the ...Proton-coupled Oligopeptide Transporters (POTs) of the Major Facilitator Superfamily (MFS) mediate the uptake of short di- and tripeptides in all phyla of life. POTs are thought to constitute the most promiscuous class of MFS transporters, with the potential to transport more than 8400 unique substrates. Over the past two decades, transport assays and biophysical studies have shown that various orthologues and paralogues display differences in substrate selectivity. The genome codes for four different POTs, known as Di- and tripeptide permeases A-D (DtpA-D). DtpC was shown previously to favor positively charged peptides as substrates. In this study, we describe, how we determined the structure of the 53 kDa DtpC by cryogenic electron microscopy (cryo-EM), and provide structural insights into the ligand specificity of this atypical POT. We collected and analyzed data on the transporter fused to split superfolder GFP (split sfGFP), in complex with a 52 kDa Pro-macrobody and with a 13 kDa nanobody. The latter sample was more stable, rigid and a significant fraction dimeric, allowing us to reconstruct a 3D volume of DtpC at a resolution of 2.7 Å. This work provides a molecular explanation for the selectivity of DtpC, and highlights the value of small and rigid fiducial markers such as nanobodies for structure determination of low molecular weight integral membrane proteins lacking soluble domains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14618.map.gz emd_14618.map.gz | 5.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14618-v30.xml emd-14618-v30.xml emd-14618.xml emd-14618.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14618_fsc.xml emd_14618_fsc.xml | 13.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_14618.png emd_14618.png | 180.7 KB | ||
| Masks |  emd_14618_msk_1.map emd_14618_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14618.cif.gz emd-14618.cif.gz | 7.1 KB | ||
| Others |  emd_14618_additional_1.map.gz emd_14618_additional_1.map.gz emd_14618_additional_2.map.gz emd_14618_additional_2.map.gz emd_14618_additional_3.map.gz emd_14618_additional_3.map.gz emd_14618_half_map_1.map.gz emd_14618_half_map_1.map.gz emd_14618_half_map_2.map.gz emd_14618_half_map_2.map.gz | 10 MB 227 MB 227 MB 226.3 MB 226.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14618 http://ftp.pdbj.org/pub/emdb/structures/EMD-14618 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14618 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14618 | HTTPS FTP |
-Validation report
| Summary document |  emd_14618_validation.pdf.gz emd_14618_validation.pdf.gz | 900.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14618_full_validation.pdf.gz emd_14618_full_validation.pdf.gz | 899.8 KB | Display | |
| Data in XML |  emd_14618_validation.xml.gz emd_14618_validation.xml.gz | 19.3 KB | Display | |
| Data in CIF |  emd_14618_validation.cif.gz emd_14618_validation.cif.gz | 24.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14618 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14618 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14618 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14618 | HTTPS FTP |
-Related structure data
| Related structure data |  7zc2MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14618.map.gz / Format: CCP4 / Size: 5.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14618.map.gz / Format: CCP4 / Size: 5.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed map. Focused ref on one copy of DtpC. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
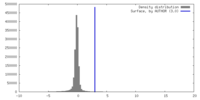
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14618_msk_1.map emd_14618_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
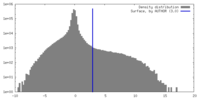
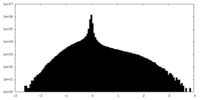
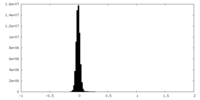
| Density Histograms |
-Additional map: Post-processed map. Dimeric reconstruction in complex with nanobody...
| File | emd_14618_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
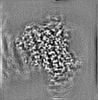
| Annotation | Post-processed map. Dimeric reconstruction in complex with nanobody 26. | ||||||||||||
| Projections & Slices |
| ||||||||||||
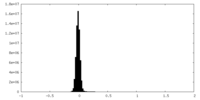
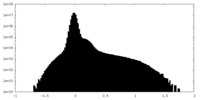
| Density Histograms |
-Additional map: Half map 1. Dimeric reconstruction in complex with nanobody 26.
| File | emd_14618_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1. Dimeric reconstruction in complex with nanobody 26. | ||||||||||||
| Projections & Slices |
| ||||||||||||
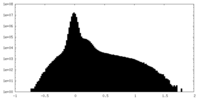
| Density Histograms |
-Additional map: Half map 2. Dimeric reconstruction in complex with nanobody 26.
| File | emd_14618_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2. Dimeric reconstruction in complex with nanobody 26. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1. Focused ref on one copy of DtpC.
| File | emd_14618_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1. Focused ref on one copy of DtpC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
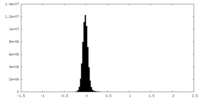
| Density Histograms |
-Half map: Half map 2. Focused ref on one copy of DtpC.
| File | emd_14618_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2. Focused ref on one copy of DtpC. | ||||||||||||
| Projections & Slices |
| ||||||||||||
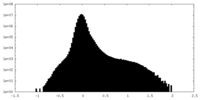
| Density Histograms |
- Sample components
Sample components
-Entire : Dipeptide and tripeptide permease C (DtpC)
| Entire | Name: Dipeptide and tripeptide permease C (DtpC) |
|---|---|
| Components |
|
-Supramolecule #1: Dipeptide and tripeptide permease C (DtpC)
| Supramolecule | Name: Dipeptide and tripeptide permease C (DtpC) / type: cell / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Amino acid/peptide transporter
| Macromolecule | Name: Amino acid/peptide transporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.090273 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTPSQPRAI YYIVAIQIWE YFSFYGMRAL LILYLTHQLG FDDNHAISLF SAYASLVYVT PILGGWLADR LLGNRTAVIA GALLMTLGH VVLGIDTNST FSLYLALAII ICGYGLFKSN ISCLLGELYD ENDHRRDGGF SLLYAAGNIG SIAAPIACGL A AQWYGWHV ...String: MKTPSQPRAI YYIVAIQIWE YFSFYGMRAL LILYLTHQLG FDDNHAISLF SAYASLVYVT PILGGWLADR LLGNRTAVIA GALLMTLGH VVLGIDTNST FSLYLALAII ICGYGLFKSN ISCLLGELYD ENDHRRDGGF SLLYAAGNIG SIAAPIACGL A AQWYGWHV GFALAGGGMF IGLLIFLSGH RHFQSTRSMD KKALTSVKFA LPVWSWLVVM LCLAPVFFTL LLENDWSGYL LA IVCLIAA QIIARMMIKF PEHRRALWQI VLLMFVGTLF WVLAQQGGST ISLFIDRFVN RQAFNIEVPT ALFQSVNAIA VML AGVVLA WLASPESRGN STLRVWLKFA FGLLLMACGF MLLAFDARHA AADGQASMGV MISGLALMGF AELFIDPVAI AQIT RLKMS GVLTGIYMLA TGAVANWLAG VVAQQTTESQ ISGMAIAAYQ RFFSQMGEWT LACVAIIVVL AFATRFLFST PTNMI QESN D UniProtKB: Peptide permease |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. |
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 24333 / Average electron dose: 75.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)