[English] 日本語
 Yorodumi
Yorodumi- EMDB-1440: The subnanometer resolution structure of the glutamate synthase 1... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1440 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
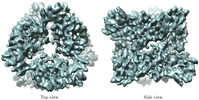
| Title | The subnanometer resolution structure of the glutamate synthase 1.2-MDa hexamer by cryoelectron microscopy and its oligomerization behavior in solution: functional implications. | |||||||||
 Map data Map data | This is the 3D map file of Azospirillum brasilense NADPH-GltS (glutamate synthase) | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationglutamate synthase (NADPH) / glutamate synthase (NADPH) activity / L-glutamate biosynthetic process / ammonia assimilation cycle / 3 iron, 4 sulfur cluster binding / 4 iron, 4 sulfur cluster binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Azospirillum brasilense (bacteria) Azospirillum brasilense (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 9.5 Å | |||||||||
 Authors Authors | Cottevieille M / Larquet E / Jonic S / Petoukhov MV / Caprini G / Paravisi S / Svergun DI / Vanoni MA / Boisset N | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2008 Journal: J Biol Chem / Year: 2008Title: The subnanometer resolution structure of the glutamate synthase 1.2-MDa hexamer by cryoelectron microscopy and its oligomerization behavior in solution: functional implications. Authors: Magali Cottevieille / Eric Larquet / Slavica Jonic / Maxim V Petoukhov / Gianluca Caprini / Stefano Paravisi / Dmitri I Svergun / Maria A Vanoni / Nicolas Boisset /  Abstract: The three-dimensional structure of the hexameric (alphabeta)(6) 1.2-MDa complex formed by glutamate synthase has been determined at subnanometric resolution by combining cryoelectron microscopy, ...The three-dimensional structure of the hexameric (alphabeta)(6) 1.2-MDa complex formed by glutamate synthase has been determined at subnanometric resolution by combining cryoelectron microscopy, small angle x-ray scattering, and molecular modeling, providing for the first time a molecular model of this complex iron-sulfur flavoprotein. In the hexameric species, interprotomeric alpha-alpha and alpha-beta contacts are mediated by the C-terminal domain of the alpha subunit, which is based on a beta helical fold so far unique to glutamate synthases. The alphabeta protomer extracted from the hexameric model is fully consistent with it being the minimal catalytically active form of the enzyme. The structure clarifies the electron transfer pathway from the FAD cofactor on the beta subunit, to the FMN on the alpha subunit, through the low potential [4Fe-4S](1+/2+) centers on the beta subunit and the [3Fe-4S](0/1+) cluster on the alpha subunit. The (alphabeta)(6) hexamer exhibits a concentration-dependent equilibrium with alphabeta monomers and (alphabeta)(2) dimers, in solution, the hexamer being destabilized by high ionic strength and, to a lower extent, by the reaction product NADP(+). Hexamerization seems to decrease the catalytic efficiency of the alphabeta protomer only 3-fold by increasing the K(m) values measured for l-Gln and 2-OG. However, it cannot be ruled out that the (alphabeta)(6) hexamer acts as a scaffold for the assembly of multienzymatic complexes of nitrogen metabolism or that it provides a means to regulate the activity of the enzyme through an as yet unknown ligand. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1440.map.gz emd_1440.map.gz | 25.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1440-v30.xml emd-1440-v30.xml emd-1440.xml emd-1440.xml | 12.3 KB 12.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_1440_fsc.xml emd_1440_fsc.xml | 7.3 KB | Display |  FSC data file FSC data file |
| Images |  1440.gif 1440.gif emd_1440.tif emd_1440.tif emd_1440_1.tif emd_1440_1.tif | 21.9 KB 487 KB 308.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1440 http://ftp.pdbj.org/pub/emdb/structures/EMD-1440 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1440 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1440 | HTTPS FTP |
-Related structure data
| Related structure data |  2vdcMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1440.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1440.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the 3D map file of Azospirillum brasilense NADPH-GltS (glutamate synthase) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.5875 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Azospirillum brasilense NADPH-GltS
| Entire | Name: Azospirillum brasilense NADPH-GltS |
|---|---|
| Components |
|
-Supramolecule #1000: Azospirillum brasilense NADPH-GltS
| Supramolecule | Name: Azospirillum brasilense NADPH-GltS / type: sample / ID: 1000 / Oligomeric state: Glutamate synthase is a heterohexamer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 1.2 MDa / Theoretical: 1.29 MDa / Method: small-angle X-ray scattering |
-Macromolecule #1: NADPH-glutamate synthase
| Macromolecule | Name: NADPH-glutamate synthase / type: protein_or_peptide / ID: 1 / Name.synonym: bacterial glutamate synthase Details: Additional InterPro entries: IPR006982 Glutamate synthase, central-N IPR012220 Glutamate synthase, eukaryotic IPR012061 Glutamate synthase, large subunit region 3, putative IPR012375 ...Details: Additional InterPro entries: IPR006982 Glutamate synthase, central-N IPR012220 Glutamate synthase, eukaryotic IPR012061 Glutamate synthase, large subunit region 3, putative IPR012375 Glutamate synthase, large subunit region 1 amidotransferase, putative IPR002932 Glutamate synthase, central-C IPR002489 Glutamate synthase, alpha subunit, C-terminal IPR014666 Ferredoxin-dependent glutamate synthase IPR012075 Glutamate synthase, large subunit region 1 and 3, putative IPR006006 Glutamate synthase, NADH/NADPH, small subunit 2 IPR006005 Glutamate synthase, NADH/NADPH, small subunit 1 Number of copies: 6 / Oligomeric state: (alpha)6 (beta)6 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Azospirillum brasilense (bacteria) / Strain: Sp7 / Location in cell: cytoplasm Azospirillum brasilense (bacteria) / Strain: Sp7 / Location in cell: cytoplasm |
| Molecular weight | Experimental: 1.29 MDa / Theoretical: 1.2 MDa |
| Recombinant expression | Organism:  |
| Sequence | GO: glutamate synthase (NADPH) activity / InterPro: Glutamate synthase, central-N |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 9.25 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 25 mM Hepes/KOH, 1 mM EDTA, 1 mM DTT |
| Staining | Type: NEGATIVE Details: CRYOEM : 4 microL were applied on a 200 mesh copper grid, coated with a thin holey carbon film. After blotting the excess of solution with Whatman paper, the grid was rapidly plunged into liquid ethane |
| Grid | Details: 200 mesh copper grid, coated with a thin holey |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: manual plunger / Method: Manual single-sided blotting |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010UHR |
|---|---|
| Temperature | Min: 91.15 K / Max: 93.15 K / Average: 93.15 K |
| Details | low-dose illumination |
| Date | May 10, 2005 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 1.59 µm / Number real images: 151 / Average electron dose: 10 e/Å2 / Details: Scanner model : Nikon Coolscan 8000ED / Bits/pixel: 8 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.5 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.7 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Gatan / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
-Atomic model buiding 1
| Software | Name:  Chimera Chimera |
|---|---|
| Details | Protocol: Rigid Body. One alpha dimer (1EA0) and one beta model (homology modelling) were separately fitted by manual docking using Chimera software. D3 symmetry was used to reconstruct the whole complex. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-2vdc: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















