-Search query
-Search result
Showing 1 - 50 of 111 items for (author: stagg & s)
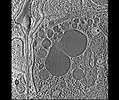
EMDB-28138: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
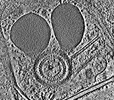
EMDB-28141: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
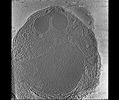
EMDB-28142: 
3D Reconstruction of Plasmodium falciparum (3D7) free merozoite

EMDB-28125: 
Plasmodium falciparum merozoites apical 2-ring units

EMDB-28126: 
Toxoplasma apical rings

EMDB-27593: 
The structure of S. epidermidis Cas10-Csm bound to target RNA
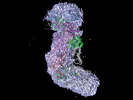
EMDB-27762: 
318 kDa Cas10-Csm effector complex bound to cognate target RNA

PDB-8do6: 
The structure of S. epidermidis Cas10-Csm bound to target RNA

EMDB-28147: 
Rabbit muscle aldolase determined using single-particle cryo-EM with Apollo camera.

EMDB-28259: 
Mouse apoferritin heavy chain with zinc determined using single-particle cryo-EM with Apollo camera.

EMDB-28269: 
Mouse apoferritin heavy chain without zinc determined using single-particle cryo-EM with Apollo camera.

PDB-8ehg: 
Rabbit muscle aldolase determined using single-particle cryo-EM with Apollo camera.

PDB-8emq: 
Mouse apoferritin heavy chain with zinc determined using single-particle cryo-EM with Apollo camera.

PDB-8en7: 
Mouse apoferritin heavy chain without zinc determined using single-particle cryo-EM with Apollo camera.

EMDB-26172: 
Global average of aligned subtomograms of AAV2 bound with PKD1-2

EMDB-26173: 
First class of AAV2 bound with PKD1-2 revealed by classification of aligned subtomgrams

EMDB-26174: 
Second class of AAV2 bound with PKD1-2 revealed by classification of aligned subtomgrams

EMDB-26175: 
Third class of AAV2 bound with PKD1-2 revealed by classification of aligned subtomgrams

EMDB-26176: 
Fourth class of AAV2 bound with PKD1-2 revealed by classification of aligned subtomgrams

EMDB-26177: 
SPR reconstruction of AAV5 bound with PKD1-2

EMDB-26182: 
Global average of aligned subtomograms of AAV5 bound with PKD1-2

EMDB-26186: 
First class of AAV5 bound with PKD1-2 revealed by classification of aligned subtomgrams

EMDB-26187: 
Second class of AAV5 bound with PKD1-2 revealed by classification of aligned subtomgrams

EMDB-26189: 
Third class of AAV5 bound with PKD1-2 revealed by classification of aligned subtomograms

EMDB-20121: 
Structure of trans-translation inhibitor bound to E. coli 70S ribosome with P site tRNA

PDB-6om6: 
Structure of trans-translation inhibitor bound to E. coli 70S ribosome with P site tRNA

EMDB-21608: 
Mini-coat geometry for a clathrin coated vesicle: clathrin-focused

EMDB-21609: 
Mini-coat geometry for a clathrin coated vesicle

EMDB-21610: 
Basket geometry for a clathrin coated vesicle

EMDB-21611: 
Asymmetric vertex of the clathrin minicoat cage, subparticle refinement of the clathrin layer

EMDB-21612: 
Football geometry for a clathrin coated vesicle

EMDB-21613: 
Tennis geometry for a clathrin coated vesicle

EMDB-21614: 
Barrel geometry for a clathrin coated vesicle

EMDB-21615: 
Pentagonal face of the mini-coat cage for a clathrin coated vesicle

EMDB-21616: 
Hexagonal face of the mini-coat cage for a clathrin coated vesicle

PDB-6wcj: 
Asymmetric vertex of the clathrin minicoat cage

EMDB-20905: 
Yeast RTP Interacts with Extended Termini of Clinet Protein Nop58p

EMDB-20155: 
Apoferritin from equine spleen at 2.9 Angstrom

EMDB-20156: 
Apoferritin from equine spleen at 3.9 Angstrom

EMDB-20157: 
Apoferritin from equine spleen at 4.3 Angstrom

EMDB-20158: 
Ribosome 70S at 2.8 Angstrom

EMDB-0553: 
Cryo-EM structure of AAV-2 in complex with AAVR PKD domains 1 and 2

PDB-6nz0: 
Cryo-EM structure of AAV-2 in complex with AAVR PKD domains 1 and 2

EMDB-0621: 
Structure of the AAV2 with its Cell Receptor, AAVR
EMDB-0622: 
Structure of the AAV2 with its Cell Receptor, AAVR

EMDB-0623: 
Structure of the AAV2 with its Cell Receptor, AAVR

EMDB-0624: 
Structure of the AAV2 with its Cell Receptor, AAVR

PDB-6ci1: 
The Structure of Full-Length Kv Beta 2.1 Determined by Cryogenic Electron Microscopy

EMDB-7305: 
The Structure of Full-Length Kv Beta 2.1 Determined by Cryogenic Electron Microscopy
EMDB-7527: 
Class 1 vertex with 2-fold symmetry extracted from SEC13/SEC31deltaC cuboctahedrons and aligned, both using localized reconstruction method.
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
