[English] 日本語
 Yorodumi
Yorodumi- EMDB-7527: Class 1 vertex with 2-fold symmetry extracted from SEC13/SEC31del... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7527 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Class 1 vertex with 2-fold symmetry extracted from SEC13/SEC31deltaC cuboctahedrons and aligned, both using localized reconstruction method. | |||||||||
 Map data Map data | Class 1 of locally reconstructed vertices of SEC13/SEC31 cages made of C-terminally truncated SEC31. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
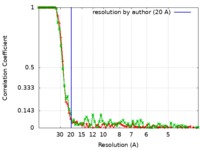
| Method | single particle reconstruction / cryo EM / Resolution: 20.0 Å | |||||||||
 Authors Authors | Paraan M / Bhattacharya N / Stagg S | |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2018 Journal: J Struct Biol / Year: 2018Title: Flexibility of the Sec13/31 cage is influenced by the Sec31 C-terminal disordered domain. Authors: Mohammadreza Paraan / Nilakshee Bhattacharya / Vladimir N Uversky / Scott M Stagg /   Abstract: In COPII mediated vesicle formation, Sec13/Sec31 heterotetramers play a role in organizing the membranes into a spherical vesicle. There they oligomerize into a cage that interacts with the other ...In COPII mediated vesicle formation, Sec13/Sec31 heterotetramers play a role in organizing the membranes into a spherical vesicle. There they oligomerize into a cage that interacts with the other COPII proteins to direct vesicle formation and concentrate cargo into a bud. In this role they must be flexible to accommodate different sizes and shapes of cargo, but also have elements that provide rigidity to help deform the membrane. Here we characterize the influence the C-terminal disordered region of Sec31 has on cage flexibility and rigidity. After deleting this region (residues 820-1220), we characterized Sec13/Sec31ΔC heterotetramers biophysically and structurally through cryo-EM. Our results show that Sec13/31ΔC self-assembles into canonical cuboctahedral cages in vitro at buffer conditions similar to wild type. The distribution of cage sizes indicated that unlike the wild type, Sec13/31ΔC cages have a more homogeneous geometry. However, the structure of cuboctahedrons exhibited more conformational heterogeneity than wild type. Through localized reconstruction of cage vertices and molecular dynamics flexible fitting we found a new hinge for the flexing of Sec31 β-propeller domain and more flexibility of the previously known hinge. Together, these results show that the C-terminal region of Sec31 regulates the flexing of other domains such that flexibility and rigidity are not compromised during transport of large and/or asymmetric cargo. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7527.map.gz emd_7527.map.gz | 9.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7527-v30.xml emd-7527-v30.xml emd-7527.xml emd-7527.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7527_fsc_1.xml emd_7527_fsc_1.xml emd_7527_fsc_2.xml emd_7527_fsc_2.xml | 7.1 KB 7.1 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_7527.png emd_7527.png | 86.6 KB | ||
| Others |  emd_7527_additional.map.gz emd_7527_additional.map.gz | 9.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7527 http://ftp.pdbj.org/pub/emdb/structures/EMD-7527 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7527 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7527 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7527.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7527.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Class 1 of locally reconstructed vertices of SEC13/SEC31 cages made of C-terminally truncated SEC31. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
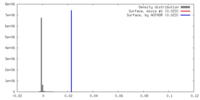
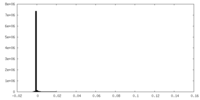
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Class 2 of locally reconstructed vertices of SEC13/SEC31...
| File | emd_7527_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Class 2 of locally reconstructed vertices of SEC13/SEC31 cages made of C-terminally truncated SEC31. | ||||||||||||
| Projections & Slices |
| ||||||||||||
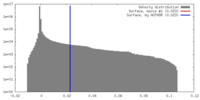
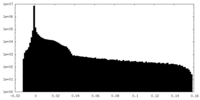
| Density Histograms |
- Sample components
Sample components
-Entire : Assembly of 24 SEC13/SEC31deltaC heterotetramers into a cuboctahe...
| Entire | Name: Assembly of 24 SEC13/SEC31deltaC heterotetramers into a cuboctahedron for which localized reconstruction reveals vertices (4 heterotetramers). |
|---|---|
| Components |
|
-Supramolecule #1: Assembly of 24 SEC13/SEC31deltaC heterotetramers into a cuboctahe...
| Supramolecule | Name: Assembly of 24 SEC13/SEC31deltaC heterotetramers into a cuboctahedron for which localized reconstruction reveals vertices (4 heterotetramers). type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Number real images: 2528 / Average exposure time: 1.08 sec. / Average electron dose: 60.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)