-Search query
-Search result
Showing 1 - 50 of 7,106 items for (author: shi & l)

EMDB-44635: 
Inactive mu opioid receptor bound to Nb6, naloxone and NAM

PDB-9bjk: 
Inactive mu opioid receptor bound to Nb6, naloxone and NAM

EMDB-38617: 
SARS-CoV-2 RBD + IMCAS-123 + IMCAS-72 Fab

EMDB-38618: 
SARS-CoV-2 RBD + IMCAS-364 + hACE2

EMDB-38619: 
SARS-CoV-2 RBD + IMCAS-364 (Local Refinement)

EMDB-38620: 
SARS-CoV-2 Omicron BA.4 RBD + IMCAS-316 + ACE2

EMDB-38621: 
SARS-CoV-2 spike + IMCAS-123

EMDB-38823: 
Cryo-EM structure of the 123-316 scDb/PT-RBD complex

PDB-8xse: 
SARS-CoV-2 RBD + IMCAS-123 + IMCAS-72 Fab

PDB-8xsf: 
SARS-CoV-2 RBD + IMCAS-364 + hACE2

PDB-8xsi: 
SARS-CoV-2 RBD + IMCAS-364 (Local Refinement)

PDB-8xsj: 
SARS-CoV-2 Omicron BA.4 RBD + IMCAS-316 + ACE2

PDB-8xsl: 
SARS-CoV-2 spike + IMCAS-123

PDB-8y0y: 
Cryo-EM structure of the 123-316 scDb/PT-RBD complex

EMDB-44395: 
Full-length cross-linked Contactin 2 (CNTN2)

EMDB-44396: 
Cross-linked Contactin 2 Ig1-Ig6

EMDB-44397: 
Full-length cross-linked Contactin 2 (FN1 apart)

EMDB-19905: 
Halobacterium salinarum archaellum filament

EMDB-19962: 
Archaellum filament from the Halobacterium salinarum deltaAgl27 strain

EMDB-36073: 
Overall structure of the LAT1-4F2hc bound with L-dopa

PDB-8j8l: 
Overall structure of the LAT1-4F2hc bound with L-dopa

EMDB-50860: 
Cryo EM map of the type 2A polymorph of alpha-synuclein at pH 7.0.

EMDB-50888: 
Cryo EM structure of the type 3B polymorph of alpha-synuclein at low pH.

PDB-9fyp: 
Cryo EM structure of the type 3B polymorph of alpha-synuclein at low pH.

EMDB-19129: 
A DNA Robotic Switch with Regulated Autonomous Display of Cytotoxic Ligand Nanopatterns

EMDB-38418: 
A neutralizing nanobody VHH60 against wt SARS-CoV-2

PDB-8xki: 
A neutralizing nanobody VHH60 against wt SARS-CoV-2

EMDB-39901: 
cryo-EM structure of the octreotide-bound SSTR5-Gi complex

EMDB-39931: 
Cryo-EM structure of the pasireotide-bound SSTR5-Gi complex

PDB-8zbe: 
cryo-EM structure of the octreotide-bound SSTR5-Gi complex

PDB-8zcj: 
Cryo-EM structure of the pasireotide-bound SSTR5-Gi complex

EMDB-18135: 
Cryo-EM structure of the methanogenic Na+ translocating N5-methyl-H4MPT:CoM methyltransferase complex

PDB-8q3v: 
Cryo-EM structure of the methanogenic Na+ translocating N5-methyl-H4MPT:CoM methyltransferase complex

EMDB-35361: 
Overall structure of the LAT1-4F2hc bound with tyrosine

PDB-8ida: 
Overall structure of the LAT1-4F2hc bound with tyrosine

EMDB-36074: 
Overall structure of the LAT1-4F2hc bound with tryptophan

PDB-8j8m: 
Overall structure of the LAT1-4F2hc bound with tryptophan

EMDB-41107: 
CryoEM structure of TR-TRAP

PDB-8t9d: 
CryoEM structure of TR-TRAP

EMDB-37105: 
ATP-bound hMRP5 outward-open
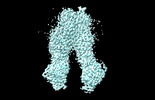
EMDB-37554: 
wt-hMRP5 inward-open
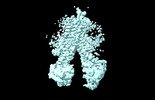
EMDB-37555: 
RD-hMRP5-inward open
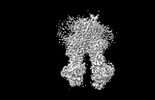
EMDB-37556: 
ND-hMRP5-inward open

EMDB-37557: 
m6-hMRP5 inward open
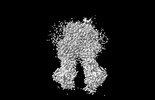
EMDB-37558: 
M5PI-bound hMRP5

PDB-8kci: 
ATP-bound hMRP5 outward-open

PDB-8wi0: 
wt-hMRP5 inward-open

PDB-8wi2: 
RD-hMRP5-inward open

PDB-8wi3: 
ND-hMRP5-inward open

PDB-8wi4: 
m6-hMRP5 inward open
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
