+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Overall structure of the LAT1-4F2hc bound with tyrosine | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / membrane protein / amino acid | |||||||||
| Function / homology |  Function and homology information Function and homology informationL-tryptophan transmembrane transport / positive regulation of L-leucine import across plasma membrane / L-tryptophan transmembrane transporter activity / cellular response to L-arginine / apical pole of neuron / tyrosine transport / L-histidine transport / amino acid transport complex / L-leucine import across plasma membrane / L-alanine transmembrane transporter activity ...L-tryptophan transmembrane transport / positive regulation of L-leucine import across plasma membrane / L-tryptophan transmembrane transporter activity / cellular response to L-arginine / apical pole of neuron / tyrosine transport / L-histidine transport / amino acid transport complex / L-leucine import across plasma membrane / L-alanine transmembrane transporter activity / L-alanine import across plasma membrane / Defective SLC7A7 causes lysinuric protein intolerance (LPI) / aromatic amino acid transmembrane transporter activity / phenylalanine transport / methionine transport / L-leucine transmembrane transporter activity / amino acid transmembrane transport / thyroid hormone transmembrane transporter activity / isoleucine transport / valine transport / proline transport / alanine transport / L-amino acid transmembrane transporter activity / L-leucine transport / thyroid hormone transport / negative regulation of vascular associated smooth muscle cell apoptotic process / neutral amino acid transport / positive regulation of cytokine production involved in immune response / amino acid import across plasma membrane / external side of apical plasma membrane / neutral L-amino acid transmembrane transporter activity / Tryptophan catabolism / exogenous protein binding / Amino acid transport across the plasma membrane / amino acid transmembrane transporter activity / anchoring junction / antiporter activity / Basigin interactions / xenobiotic transport / response to muscle activity / microvillus membrane / positive regulation of interleukin-4 production / response to exogenous dsRNA / positive regulation of interleukin-17 production / amino acid transport / tryptophan transport / positive regulation of glial cell proliferation / response to hyperoxia / transport across blood-brain barrier / cellular response to glucose starvation / liver regeneration / negative regulation of autophagy / basal plasma membrane / peptide antigen binding / positive regulation of type II interferon production / calcium ion transport / melanosome / double-stranded RNA binding / cellular response to lipopolysaccharide / virus receptor activity / basolateral plasma membrane / carbohydrate metabolic process / apical plasma membrane / cadherin binding / protein heterodimerization activity / negative regulation of gene expression / lysosomal membrane / intracellular membrane-bounded organelle / synapse / symbiont entry into host cell / cell surface / protein homodimerization activity / RNA binding / extracellular exosome / nucleoplasm / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Yan RH / Li YN / Shi TH | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2025 Journal: J Biol Chem / Year: 2025Title: Structural insights into the substrate transport mechanism of the amino acid transporter complex. Authors: Haonan Yang / Tianhao Shi / Jing Dong / Ting Zhang / Yaning Li / Yingying Guo / Yafei Yuan / Liuqing Yang / Jin-Tang Dong / Renhong Yan /  Abstract: The -type amino acid transporter 1 (LAT1), in complex with its ancillary protein 4F2hc, mediates the sodium-independent antiport of large neutral amino acids across the plasma membrane. LAT1 ...The -type amino acid transporter 1 (LAT1), in complex with its ancillary protein 4F2hc, mediates the sodium-independent antiport of large neutral amino acids across the plasma membrane. LAT1 preferentially transports substrates, such as -leucine, -tyrosine, and -tryptophan, thyroid hormones, and drugs like 3,4-dihydroxyphenylalanine. Its pivotal role in cancer development and progression has established LAT1 as a promising therapeutic target. While prior studies have resolved the LAT1-4F2hc architecture and inhibitor interactions, the molecular basis of LAT1 substrate selectivity remains elusive. Here, we present the cryo-EM structures of LAT1-4F2hc bound to -tyrosine, -tryptophan, -leucine, and 3,4-dihydroxyphenylalanine, revealing distinct substrate binding modes. Comparative structural analysis highlights differences between LAT1 and LAT2 in substrate coordination, driven by key residues near the binding pocket that influence transport efficiency. These findings advance our mechanistic understanding of the LAT1-4F2hc complex and provide valuable insights for structure-based drug design targeting LAT1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35361.map.gz emd_35361.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35361-v30.xml emd-35361-v30.xml emd-35361.xml emd-35361.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35361.png emd_35361.png | 87.8 KB | ||
| Filedesc metadata |  emd-35361.cif.gz emd-35361.cif.gz | 7 KB | ||
| Others |  emd_35361_half_map_1.map.gz emd_35361_half_map_1.map.gz emd_35361_half_map_2.map.gz emd_35361_half_map_2.map.gz | 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35361 http://ftp.pdbj.org/pub/emdb/structures/EMD-35361 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35361 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35361 | HTTPS FTP |
-Validation report
| Summary document |  emd_35361_validation.pdf.gz emd_35361_validation.pdf.gz | 795.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35361_full_validation.pdf.gz emd_35361_full_validation.pdf.gz | 795.2 KB | Display | |
| Data in XML |  emd_35361_validation.xml.gz emd_35361_validation.xml.gz | 12.1 KB | Display | |
| Data in CIF |  emd_35361_validation.cif.gz emd_35361_validation.cif.gz | 14.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35361 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35361 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35361 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35361 | HTTPS FTP |
-Related structure data
| Related structure data |  8idaMC  8j8lC  8j8mC  8x0wC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35361.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35361.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.087 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35361_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
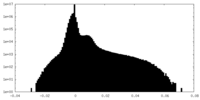
| Projections & Slices |
| ||||||||||||
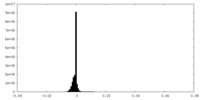
| Density Histograms |
-Half map: #1
| File | emd_35361_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
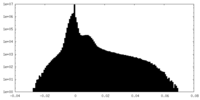
| Projections & Slices |
| ||||||||||||
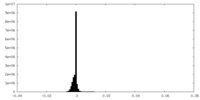
| Density Histograms |
- Sample components
Sample components
-Entire : Overall structure of the LAT1-4F2hc bound with tyrosine
| Entire | Name: Overall structure of the LAT1-4F2hc bound with tyrosine |
|---|---|
| Components |
|
-Supramolecule #1: Overall structure of the LAT1-4F2hc bound with tyrosine
| Supramolecule | Name: Overall structure of the LAT1-4F2hc bound with tyrosine type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: 4F2 cell-surface antigen heavy chain
| Macromolecule | Name: 4F2 cell-surface antigen heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 70.160828 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAHHHHHHHH HHSGRELQPP EASIAVVSIP RQLPGSHSEA GVQGLSAGDD SETGSDCVTQ AGLQLLASSD PPALASKNAE VTVETGFHH VSQADIEFLT SIDPTASASG SAGITGTMSQ DTEVDMKEVE LNELEPEKQP MNAASGAAMS LAGAEKNGLV K IKVAEDEA ...String: MAHHHHHHHH HHSGRELQPP EASIAVVSIP RQLPGSHSEA GVQGLSAGDD SETGSDCVTQ AGLQLLASSD PPALASKNAE VTVETGFHH VSQADIEFLT SIDPTASASG SAGITGTMSQ DTEVDMKEVE LNELEPEKQP MNAASGAAMS LAGAEKNGLV K IKVAEDEA EAAAAAKFTG LSKEELLKVA GSPGWVRTRW ALLLLFWLGW LGMLAGAVVI IVRAPRCREL PAQKWWHTGA LY RIGDLQA FQGHGAGNLA GLKGRLDYLS SLKVKGLVLG PIHKNQKDDV AQTDLLQIDP NFGSKEDFDS LLQSAKKKSI RVI LDLTPN YRGENSWFST QVDTVATKVK DALEFWLQAG VDGFQVRDIE NLKDASSFLA EWQNITKGFS EDRLLIAGTN SSDL QQILS LLESNKDLLL TSSYLSDSGS TGEHTKSLVT QYLNATGNRW CSWSLSQARL LTSFLPAQLL RLYQLMLFTL PGTPV FSYG DEIGLDAAAL PGQPMEAPVM LWDESSFPDI PGAVSANMTV KGQSEDPGSL LSLFRRLSDQ RSKERSLLHG DFHAFS AGP GLFSYIRHWD QNERFLVVLN FGDVGLSAGL QASDLPASAS LPAKADLLLS TQPGREEGSP LELERLKLEP HEGLLLR FP YAALE UniProtKB: Amino acid transporter heavy chain SLC3A2 |
-Macromolecule #2: Large neutral amino acids transporter small subunit 1
| Macromolecule | Name: Large neutral amino acids transporter small subunit 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.186895 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MADYKDDDDK SGPDEVDASG RAGAGPKRRA LAAPAAEEKE EAREKMLAAK SADGSAPAGE GEGVTLQRNI TLLNGVAIIV GTIIGSGIF VTPTGVLKEA GSPGLALVVW AACGVFSIVG ALCYAELGTT ISKSGGDYAY MLEVYGSLPA FLKLWIELLI I RPSSQYIV ...String: MADYKDDDDK SGPDEVDASG RAGAGPKRRA LAAPAAEEKE EAREKMLAAK SADGSAPAGE GEGVTLQRNI TLLNGVAIIV GTIIGSGIF VTPTGVLKEA GSPGLALVVW AACGVFSIVG ALCYAELGTT ISKSGGDYAY MLEVYGSLPA FLKLWIELLI I RPSSQYIV ALVFATYLLK PLFPTCPVPE EAAKLVACLC VLLLTAVNCY SVKAATRVQD AFAAAKLLAL ALIILLGFVQ IG KGDVSNL DPNFSFEGTK LDVGNIVLAL YSGLFAYGGW NYLNFVTEEM INPYRNLPLA IIISLPIVTL VYVLTNLAYF TTL STEQML SSEAVAVDFG NYHLGVMSWI IPVFVGLSCF GSVNGSLFTS SRLFFVGSRE GHLPSILSMI HPQLLTPVPS LVFT CVMTL LYAFSKDIFS VINFFSFFNW LCVALAIIGM IWLRHRKPEL ERPIKVNLAL PVFFILACLF LIAVSFWKTP VECGI GFTI ILSGLPVYFF GVWWKNKPKW LLQGIFSTTV LCQKLMQVVP QET UniProtKB: Large neutral amino acids transporter small subunit 1 |
-Macromolecule #4: TYROSINE
| Macromolecule | Name: TYROSINE / type: ligand / ID: 4 / Number of copies: 1 / Formula: TYR |
|---|---|
| Molecular weight | Theoretical: 181.189 Da |
| Chemical component information | 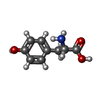 ChemComp-TYR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)