-Search query
-Search result
Showing 1 - 50 of 147 items for (author: shek & r)

EMDB-41260: 
SARS-CoV-2 BA.1 S-6P-no-RBD

EMDB-18402: 
cryo-EM structure of apo-TcdB

EMDB-18403: 
cryo-EM map of apo Clostridioides difficile toxin B

EMDB-18409: 
cryo-EM structure of TcdB-FZD7

EMDB-18410: 
cryo-EM structure of TcdB-FZD7

EMDB-18411: 
cryo-EM structure of TcdB-FZD7
EMDB-18639: 
Locally refined SARS-CoV-2 BA-2.86 Spike receptor binding domain (RBD) complexed with angiotensin converting enzyme 2 (ACE2)

EMDB-18649: 
Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab

EMDB-19002: 
XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein

PDB-8qsq: 
Locally refined SARS-CoV-2 BA-2.86 Spike receptor binding domain (RBD) complexed with angiotensin converting enzyme 2 (ACE2)

PDB-8qtd: 
Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab

PDB-8r8k: 
XBB-4 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein

EMDB-18373: 
cryo-EM structure of apo Clostridioides difficile toxin B

EMDB-18374: 
cryo-EM structure complex of Frizzled-7 and Clostridioides difficile toxin B

PDB-8qen: 
cryo-EM structure of apo Clostridioides difficile toxin B

PDB-8qeo: 
cryo-EM structure complex of Frizzled-7 and Clostridioides difficile toxin B

EMDB-35304: 
Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3A) complex

EMDB-35306: 
Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3A-N71A) complex

EMDB-35310: 
Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3D) complex

PDB-8iaj: 
Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3A) complex

PDB-8iak: 
Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3A-N71A) complex

PDB-8iam: 
Cryo-EM structure of the yeast SPT-ORM2 (ORM2-S3D) complex

EMDB-28965: 
Glutamine synthetase from Pseudomonas aeruginosa, filament double-unit in compressed conformation

PDB-8fbp: 
Glutamine synthetase from Pseudomonas aeruginosa, filament double-unit in compressed conformation

EMDB-16569: 
PfRH5-PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to antibody Cy.003
EMDB-16570: 
PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to antibody Cy.003

EMDB-16635: 
PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to antibody Cy.003 - maps for local refinement on PfRIPR

EMDB-16636: 
PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to antibody Cy.003 consensus maps

EMDB-16637: 
PfRH5-PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to antibody Cy.003 - consensus map

EMDB-16638: 
PfRH5-PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to antibody Cy.003 - local refinement on PfRH5

EMDB-16639: 
PfRH5-PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to antibody Cy.003 - local refinement on PfRIPR

EMDB-16640: 
PfRH5-PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to antibody Cy.003 - map with additional PfRIPR tail density

PDB-8cdd: 
PfRH5-PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to antibody Cy.003

PDB-8cde: 
PfCyRPA-PfRIPR complex from Plasmodium falciparum bound to antibody Cy.003

EMDB-40671: 
Cryo-EM structure of Enoyl-CoA hydratase from Mycobacterium smegmatis

PDB-8soy: 
Cryo-EM structure of Enoyl-CoA hydratase from Mycobacterium smegmatis

EMDB-33873: 
Cryo-EM structure of the dimeric atSPT-ORM1 complex

EMDB-33874: 
Cryo-EM structure of the monomeric atSPT-ORM1 complex
EMDB-33875: 
Cryo-EM structure of the monomeric atSPT-ORM1 (ORM1-N17A) complex

EMDB-33876: 
Cryo-EM structure of the monomeric atSPT-ORM1 (LCB2a-deltaN5) complex

PDB-7yjk: 
Cryo-EM structure of the dimeric atSPT-ORM1 complex

PDB-7yjm: 
Cryo-EM structure of the monomeric atSPT-ORM1 complex

PDB-7yjn: 
Cryo-EM structure of the monomeric atSPT-ORM1 (ORM1-N17A) complex

PDB-7yjo: 
Cryo-EM structure of the monomeric atSPT-ORM1 (LCB2a-deltaN5) complex
EMDB-26945: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-21
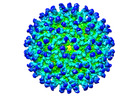
EMDB-26946: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-94
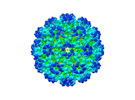
EMDB-26947: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a potently neutralizing human antibody IgG-106

PDB-7v0n: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-21

PDB-7v0o: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-94

PDB-7v0p: 
Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a potently neutralizing human antibody IgG-106
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
