+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the dimeric atSPT-ORM1 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ceramide / TRANSFERASE-inhibitor complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of sphingolipid biosynthetic process / multidimensional cell growth / leaf senescence / intracellular sphingolipid homeostasis / serine palmitoyltransferase complex / serine C-palmitoyltransferase activity / serine C-palmitoyltransferase / pollen development / photomorphogenesis / regulation of programmed cell death ...regulation of sphingolipid biosynthetic process / multidimensional cell growth / leaf senescence / intracellular sphingolipid homeostasis / serine palmitoyltransferase complex / serine C-palmitoyltransferase activity / serine C-palmitoyltransferase / pollen development / photomorphogenesis / regulation of programmed cell death / ceramide metabolic process / sphingosine biosynthetic process / embryo development ending in seed dormancy / sphingolipid biosynthetic process / vacuole / response to endoplasmic reticulum stress / pyridoxal phosphate binding / response to oxidative stress / defense response to bacterium / endoplasmic reticulum membrane / protein-containing complex binding / endoplasmic reticulum / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Xie T / Liu P / Gong X | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Mechanism of sphingolipid homeostasis revealed by structural analysis of SPT-ORM1 complex. Authors: Peng Liu / Tian Xie / Xinyue Wu / Gongshe Han / Sita D Gupta / Zike Zhang / Jian Yue / Feitong Dong / Kenneth Gable / Somashekarappa Niranjanakumari / Wanyuan Li / Lin Wang / Wenchen Liu / ...Authors: Peng Liu / Tian Xie / Xinyue Wu / Gongshe Han / Sita D Gupta / Zike Zhang / Jian Yue / Feitong Dong / Kenneth Gable / Somashekarappa Niranjanakumari / Wanyuan Li / Lin Wang / Wenchen Liu / Ruifeng Yao / Edgar B Cahoon / Teresa M Dunn / Xin Gong /   Abstract: The serine palmitoyltransferase (SPT) complex catalyzes the first and rate-limiting step in sphingolipid biosynthesis in all eukaryotes. ORM/ORMDL proteins are negative regulators of SPT that respond ...The serine palmitoyltransferase (SPT) complex catalyzes the first and rate-limiting step in sphingolipid biosynthesis in all eukaryotes. ORM/ORMDL proteins are negative regulators of SPT that respond to cellular sphingolipid levels. However, the molecular basis underlying ORM/ORMDL-dependent homeostatic regulation of SPT is not well understood. We determined the cryo-electron microscopy structure of SPT-ORM1 complex, composed of LCB1, LCB2a, SPTssa, and ORM1, in an inhibited state. A ceramide molecule is sandwiched between ORM1 and LCB2a in the cytosolic membrane leaflet. Ceramide binding is critical for the ORM1-dependent SPT repression, and dihydroceramides and phytoceramides differentially affect this repression. A hybrid β sheet, formed by the amino termini of ORM1 and LCB2a and induced by ceramide binding, stabilizes the amino terminus of ORM1 in an inhibitory conformation. Our findings provide mechanistic insights into sphingolipid homeostatic regulation via the binding of ceramide to the SPT-ORM/ORMDL complex that may have implications for plant-specific processes such as the hypersensitive response for microbial pathogen resistance. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33873.map.gz emd_33873.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33873-v30.xml emd-33873-v30.xml emd-33873.xml emd-33873.xml | 25 KB 25 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33873.png emd_33873.png | 55.3 KB | ||
| Filedesc metadata |  emd-33873.cif.gz emd-33873.cif.gz | 7.6 KB | ||
| Others |  emd_33873_half_map_1.map.gz emd_33873_half_map_1.map.gz emd_33873_half_map_2.map.gz emd_33873_half_map_2.map.gz | 48.6 MB 48.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33873 http://ftp.pdbj.org/pub/emdb/structures/EMD-33873 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33873 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33873 | HTTPS FTP |
-Related structure data
| Related structure data |  7yjkMC  7yjmC  7yjnC  7yjoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33873.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33873.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
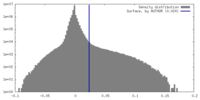
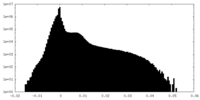
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33873_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
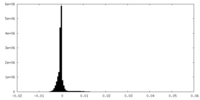
| Density Histograms |
-Half map: #1
| File | emd_33873_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
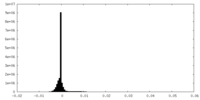
| Density Histograms |
- Sample components
Sample components
-Entire : SPT-ORM1 complex
| Entire | Name: SPT-ORM1 complex |
|---|---|
| Components |
|
-Supramolecule #1: SPT-ORM1 complex
| Supramolecule | Name: SPT-ORM1 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Long chain base biosynthesis protein 1
| Macromolecule | Name: Long chain base biosynthesis protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: serine C-palmitoyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.207453 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASNLVEMFN AALNWVTMIL ESPSARVVLF GVPIRGHFFV EGLLGVVIII LLTRKSYKPP KRPLTEQEID ELCDEWVPEP LIPPITEDM KHEPPVLESA AGPHTTVNGK DVVNFASANY LGLIGHEKLL ESCTSALEKY GVGSCGPRGF YGTIDVHLDC E TRISKFLG ...String: MASNLVEMFN AALNWVTMIL ESPSARVVLF GVPIRGHFFV EGLLGVVIII LLTRKSYKPP KRPLTEQEID ELCDEWVPEP LIPPITEDM KHEPPVLESA AGPHTTVNGK DVVNFASANY LGLIGHEKLL ESCTSALEKY GVGSCGPRGF YGTIDVHLDC E TRISKFLG TPDSILYSYG LSTMFSTIPC FCKKGDVIVA DEGVHWGIQN GLQLSRSTIV YFKHNDMESL RITLEKIMTK YK RSKNLRR YIVAEAVYQN SGQIAPLDEI VKLKEKYRFR VILDESNSFG VLGRSGRGLA EHHSVPIEKI DVVTAAMGHA LAT EGGFCT GNARIIDYQR LSSSGYVFSA SLPPYLASAA ITAIDVIDQN PDMLVKLKQN VALLWKGLSD IKGMSLTSNR ESPI VFLKL EKSSGSAKDD LLLLEKMADR ALKEDSLLVV SSKRSFLDKC RLPVGIKLYV SAGHSESDLL KASESLKRLA SELLL KS UniProtKB: Long chain base biosynthesis protein 1 |
-Macromolecule #2: Long chain base biosynthesis protein 2a
| Macromolecule | Name: Long chain base biosynthesis protein 2a / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: serine C-palmitoyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 54.587574 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MITIPYLTAV STYFSYGLLF AFGQLRDFFR RFIDWWFTSN LQGYAPICLG HEDFYIRRLY HRIQDCFERP ISSAPDAWFD VVERYSNDN NKTLKRTTKT SRCLNLGSYN YLGFGSFDEY CTPRVIESLK KFSASTCSSR VDAGTTSVHA ELEECVTRFV G KPAAVVFG ...String: MITIPYLTAV STYFSYGLLF AFGQLRDFFR RFIDWWFTSN LQGYAPICLG HEDFYIRRLY HRIQDCFERP ISSAPDAWFD VVERYSNDN NKTLKRTTKT SRCLNLGSYN YLGFGSFDEY CTPRVIESLK KFSASTCSSR VDAGTTSVHA ELEECVTRFV G KPAAVVFG MGYATNSAII PVLIGKGGLI ISDSLNHSSI VNGARGSGAT IRVFQHNTPS HLERVLREQI AEGQPRTHRP WK KIIVVVE GIYSMEGEIC HLPEVVAICK KYKAYVYLDE AHSIGAIGKT GKGICELLGV DTADVDVMMG TFT(LLP)SFGSC GGYIAGSKEL IQYLKHQCPA HLYATSIPTP SAQQIISAIK VILGEDGSNR GAQKLARIRE NSNFFRAELQ KMGFEVLGDN DSPVMPIML YNPAKIPAFS RECLRQKVAV VVVGFPATPL LLARARICIS ASHSREDLIR ALKVISKVGD LSGIKYFPAE P KKIEQSKN DIKLD UniProtKB: Long chain base biosynthesis protein 2a |
-Macromolecule #3: ORMDL family protein
| Macromolecule | Name: ORMDL family protein / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.220266 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MANLYVKAVP PPDMNRNTEW FMYPGVWTTY MLILFFGWLV VLSVSGCSPG MAWTVVNLAH FVVTYHSFHW MKGTPFADDQ GIYNGLTWW EQMDNGQQLT RNRKFLTLVP VVLYLIASHT TDYRHPWLFL NTLAVMVLVV AKFPNMHKVR IFGINGDK UniProtKB: ORMDL family protein |
-Macromolecule #4: Transmembrane protein, putative (DUF3317)
| Macromolecule | Name: Transmembrane protein, putative (DUF3317) / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.442687 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MADYKDDDDK SGPDEVDASG RMNWVQRKIY LYNVTFGLYM LDWWERYLFN SLVVVLMWFV LYNGTRYFSE LFQRHLT UniProtKB: Transmembrane protein, putative (DUF3317) |
-Macromolecule #5: N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide
| Macromolecule | Name: N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide type: ligand / ID: 5 / Number of copies: 2 / Formula: Z1T |
|---|---|
| Molecular weight | Theoretical: 650.113 Da |
| Chemical component information |  ChemComp-Z1T: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)