-Search query
-Search result
Showing all 46 items for (author: scott & sd)

EMDB-70318: 
PV2-10D2 Complex
Method: single particle / : Waddey BT, Hafenstein SL

EMDB-70320: 
SIPV3-2E1 Complex
Method: single particle / : Waddey BT, Hafenstein SL

EMDB-70339: 
SIPV3-6B5 Complex
Method: single particle / : Waddey BT, Hafenstein SL

EMDB-70392: 
SIPV1-5E12 Complex
Method: single particle / : Waddey BT, Hafenstein SL

EMDB-70822: 
Consensus Map of PI4KA/TTC7B/FAM126A/F3IN Nanobody
Method: single particle / : Shaw AL, Suresh S, Yip CK, Burke JE

EMDB-70824: 
Local Refinement A of PI4KA/TTC7B/FAM126A/F3IN Nanobody
Method: single particle / : Shaw AL, Suresh S, Yip CK, Burke JE

EMDB-70825: 
Local Refinement B of PI4KA/TTC7B/FAM126A/F3IN nanbody
Method: single particle / : Shaw AL, Suresh S, Yip CK, Burke JE

EMDB-70826: 
Cryo-EM structure of the PI4KA complex bound to an EFR3 interfering nanobody (F3IN)
Method: single particle / : Shaw AL, Suresh S, Yip CK, Burke JE

EMDB-71704: 
HU-38 Fab with PRAME pMHC
Method: single particle / : Mortenson DE, Yu X

PDB-9pkv: 
HU-38 Fab with PRAME pMHC
Method: single particle / : Mortenson DE, Yu X

EMDB-44444: 
PaMsbA in an occluded, outward conformation
Method: single particle / : Bahramimoghaddam H, Laganowsky A

EMDB-44445: 
PaMsbA in an open, outward conformation
Method: single particle / : Bahramimoghaddam H, Laganowsky A

EMDB-43792: 
phi29 empty capsid lacking pentons and portal
Method: single particle / : Woodson ME, Morais MC, Seth S, Zhang W, Jardine PJ

EMDB-43793: 
phi29 empty capsid maturation intermediate with wild-type pRNA
Method: single particle / : Woodson ME, Morais MC, Zhang W, Jardine PJ

EMDB-41596: 
KDL bound, nucleotide-free MsbA in open, outward-facing conformation
Method: single particle / : Yang B, Zhang T, Lyu J, Laganowsky AD, Zhao M

EMDB-41597: 
Open, inward-facing MsbA structure (OIF1)
Method: single particle / : Yang B, Zhang T, Lyu J, Laganowsky AD, Zhao M

EMDB-41598: 
Open, inward-facing MsbA structure (OIF2)
Method: single particle / : Yang B, Zhang T, Lyu J, Laganowsky AD, Zhao M

EMDB-41599: 
Open, inward-facing MsbA structure (OIF4)
Method: single particle / : Yang B, Zhang T, Lyu J, Laganowsky AD, Zhao M

EMDB-41600: 
Open, inward-facing MsbA structure (OIF3)
Method: single particle / : Yang B, Zhang T, Lyu J, Laganowsky AD, Zhao M

EMDB-28820: 
phi-29 prohead MCP gp8 penton maturation intermediate, with associated scaffold gp7 tetramer
Method: single particle / : Woodson ME, Morais MC, Jardine PJ, Zhang W, Prokhorov NS

EMDB-28821: 
bacteriophage phi-29 MCP gp-8 penton in intermediate maturation state, with associated scaffold gp7 dimer
Method: single particle / : Woodson ME, Morais MC, Prokhorov NS, Zhang W, Jardine PJ

EMDB-28822: 
Phi-29 scaffolding protein bound to intermediate-state MCP
Method: single particle / : Woodson ME, Morais MC, Jardine PJ, Scott SD

EMDB-28823: 
Phi-29 partially-expanded fiberless prohead
Method: single particle / : Woodson ME, Morais MC, Scott SD, Choi KH, Jardine PJ, Zhang W

EMDB-28824: 
Phi-29 expanded, DNA-packaged fiberless prohead
Method: single particle / : Woodson ME, Morais MC, Jardine PJ, Zhang W

EMDB-27738: 
Negative stain EM map of the heterodimeric p110gamma-p84 complex
Method: single particle / : Burke JE, Dalwadi U, Rathinaswamy MK, Yip CK, Nam SE

EMDB-27627: 
Structure of p110 gamma bound to the Ras inhibitory nanobody NB7
Method: single particle / : Burke JE, Nam SE, Rathinaswamy MK, Yip CK

EMDB-23812: 
Structure of the apo phosphoinositide 3-kinase p110 gamma (PIK3CG) p101 (PIK3R5) complex
Method: single particle / : Burke JE, Dalwadi U, Rathinaswamy MK, Yip CK

EMDB-22738: 
CryoEM reconstruction of SARS-CoV-2 receptor binding domain in complex with the Fab fragment of neutralizing antibody 46
Method: single particle / : Kucharska I, Tan YZ, Benlekbir S, Rubinstein JL, Julien JP

EMDB-22739: 
CryoEM reconstruction of SARS-CoV-2 Spike in complex with the Fab fragment of neutralizing antibody 80.
Method: single particle / : Kucharska I, Tan YZ, Benlekbir S, Rubinstein JL, Julien JP

EMDB-22740: 
CryoEM reconstruction of SARS-CoV-2 Spike in complex with the Fab fragment of neutralizing antibody 298
Method: single particle / : Kucharska I, Tan YZ, Benlekbir S, Rubinstein JL, Julien JP

EMDB-22741: 
CryoEM reconstruction of SARS-CoV-2 Spike in complex with the Fab fragment of neutralizing antibody 324
Method: single particle / : Kucharska I, Tan YZ, Benlekbir S, Rubinstein JL, Julien JP

EMDB-20569: 
Monoclonal antibody 1C01 binds stem domain of influenza virus hemagglutinin
Method: single particle / : Han J, Ward A

EMDB-20570: 
Monoclonal antibody 1F03 binds head domain of influenza virus hemagglutinin
Method: single particle / : Han J, Ward A

EMDB-20571: 
Monoclonal antibody 1H09 binds head domain of influenza virus hemagglutinin
Method: single particle / : Han J, Ward A

EMDB-0339: 
Cannabinoid Receptor 1-G Protein Complex
Method: single particle / : Krishna Kumar K, Shalev-Benami M

EMDB-4089: 
The human 26S Proteasome at 6.8 Ang.
Method: single particle / : Schweitzer A, Beck F
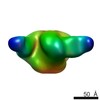
EMDB-8078: 
BG505 SOSIP.664 HIV-1 Env trimer in complex with anti-HIV CH235.12 wk323 Fab
Method: single particle / : Ozorowski G, Ward AB

EMDB-8079: 
B41 SOSIP.664 HIV-1 Env trimer in complex with anti-HIV CH235.12 wk323 Fab
Method: single particle / : Ozorowski G, Ward AB
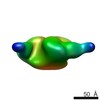
EMDB-8080: 
B41 SOSIP.664 HIV-1 Env trimer in complex with anti-HIV CH235.9 wk152 Fab
Method: single particle / : Ozorowski G, Ward AB

EMDB-8081: 
BG505 SOSIP.664 HIV-1 Env trimer in complex with anti-HIV CH235.9 wk152 Fab
Method: single particle / : Ozorowski G, Ward AB
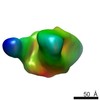
EMDB-8082: 
BG505 SOSIP.664 HIV-1 Env trimer in complex with anti-HIV CH235 wk41 Fab
Method: single particle / : Ozorowski G, Ward AB
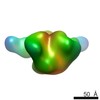
EMDB-8083: 
B41 SOSIP.664 HIV-1 Env trimer in complex with anti-HIV CH235 wk41 Fab
Method: single particle / : Ozorowski G, Ward AB

EMDB-1865: 
Structure of the yeast eisosome core component Lsp1 filament
Method: helical / : Karotki L, Huiskonen JT, Stefan CJ, Roth R, Surma MA, Aguilar PS, Krogan NJ, Emr SD, Heuser J, Gruenewald K, Walther TC

EMDB-1866: 
Structure of the yeast eisosome core component Lsp1 filament
Method: helical / : Karotki L, Huiskonen JT, Stefan CJ, Roth R, Surma MA, Aguilar PS, Krogan NJ, Emr SD, Heuser J, Gruenewald K, Walther TC

EMDB-1867: 
Structure of the yeast eisosome core component Lsp1 filament bound to a liposome membrane.
Method: helical / : Karotki L, Huiskonen JT, Stefan CJ, Roth R, Surma MA, Aguilar PS, Krogan NJ, Emr SD, Heuser J, Gruenewald K, Walther TC

EMDB-1868: 
Structure of the yeast eisosome core component Pil1 filament bound to a liposome membrane.
Method: helical / : Karotki L, Huiskonen JT, Stefan CJ, Roth R, Surma MA, Aguilar PS, Krogan NJ, Emr SD, Heuser J, Gruenewald K, Walther TC
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
