[English] 日本語
 Yorodumi
Yorodumi- EMDB-27738: Negative stain EM map of the heterodimeric p110gamma-p84 complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain EM map of the heterodimeric p110gamma-p84 complex | |||||||||
 Map data Map data | Negative stain EM ab initio model of the heterodimeric p110gamma-p84 complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PI3K / p110 / PIK3CG / phosphoinositide / PIK3R6 / p84/p87 / 3-kinase / PIP3 / IMMUNE SYSTEM / TRANSFERASE | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
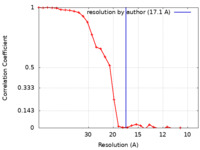
| Method | single particle reconstruction / negative staining / Resolution: 17.1 Å | |||||||||
 Authors Authors | Burke JE / Dalwadi U / Rathinaswamy MK / Yip CK / Nam SE | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2023 Journal: Cell Rep / Year: 2023Title: Molecular basis for differential activation of p101 and p84 complexes of PI3Kγ by Ras and GPCRs. Authors: Manoj K Rathinaswamy / Meredith L Jenkins / Benjamin R Duewell / Xuxiao Zhang / Noah J Harris / John T Evans / Jordan T B Stariha / Udit Dalwadi / Kaelin D Fleming / Harish Ranga-Prasad / ...Authors: Manoj K Rathinaswamy / Meredith L Jenkins / Benjamin R Duewell / Xuxiao Zhang / Noah J Harris / John T Evans / Jordan T B Stariha / Udit Dalwadi / Kaelin D Fleming / Harish Ranga-Prasad / Calvin K Yip / Roger L Williams / Scott D Hansen / John E Burke /    Abstract: Class IB phosphoinositide 3-kinase (PI3Kγ) is activated in immune cells and can form two distinct complexes (p110γ-p84 and p110γ-p101), which are differentially activated by G protein-coupled ...Class IB phosphoinositide 3-kinase (PI3Kγ) is activated in immune cells and can form two distinct complexes (p110γ-p84 and p110γ-p101), which are differentially activated by G protein-coupled receptors (GPCRs) and Ras. Using a combination of X-ray crystallography, hydrogen deuterium exchange mass spectrometry (HDX-MS), electron microscopy, molecular modeling, single-molecule imaging, and activity assays, we identify molecular differences between p110γ-p84 and p110γ-p101 that explain their differential membrane recruitment and activation by Ras and GPCRs. The p110γ-p84 complex is dynamic compared with p110γ-p101. While p110γ-p101 is robustly recruited by Gβγ subunits, p110γ-p84 is weakly recruited to membranes by Gβγ subunits alone and requires recruitment by Ras to allow for Gβγ activation. We mapped two distinct Gβγ interfaces on p101 and the p110γ helical domain, with differences in the C-terminal domain of p84 and p101 conferring sensitivity of p110γ-p101 to Gβγ activation. Overall, our work provides key insight into the molecular basis for how PI3Kγ complexes are activated. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27738.map.gz emd_27738.map.gz | 1.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27738-v30.xml emd-27738-v30.xml emd-27738.xml emd-27738.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27738_fsc.xml emd_27738_fsc.xml | 3.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_27738.png emd_27738.png | 46 KB | ||
| Filedesc metadata |  emd-27738.cif.gz emd-27738.cif.gz | 4.8 KB | ||
| Others |  emd_27738_half_map_1.map.gz emd_27738_half_map_1.map.gz emd_27738_half_map_2.map.gz emd_27738_half_map_2.map.gz | 1.3 MB 1.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27738 http://ftp.pdbj.org/pub/emdb/structures/EMD-27738 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27738 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27738 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27738.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27738.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain EM ab initio model of the heterodimeric p110gamma-p84 complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.67 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_27738_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
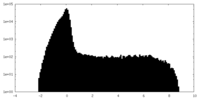
| Projections & Slices |
| ||||||||||||
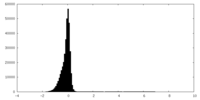
| Density Histograms |
-Half map: #2
| File | emd_27738_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
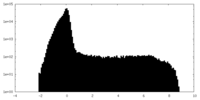
| Projections & Slices |
| ||||||||||||
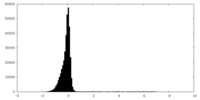
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of p110 gamma with p84
| Entire | Name: Ternary complex of p110 gamma with p84 |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of p110 gamma with p84
| Supramolecule | Name: Ternary complex of p110 gamma with p84 / type: complex / ID: 1 / Parent: 0 / Details: Both p110 and p84 subunits are from Homo sapiens |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.5 Component:
Details: Freshly prepared gel filtration buffer, filtered through 0.22 um filter and degassed | |||||||||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Formate Details: Negative stain EM samples prepares by adsorbing samples on grid for 15 second, followed by blotting of sample, 2 washes with water, 1 wash with stain, and a final 30 second soak in stain and blot. | |||||||||||||||
| Grid | Model: Homemade / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS | |||||||||||||||
| Details | Specimen was a 1:1 molar ratio of p110g to p84, purified to homogeneity by gel filtration. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 50 / Average exposure time: 1.0 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 6.3 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 49000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)