[English] 日本語
 Yorodumi
Yorodumi- EMDB-22738: CryoEM reconstruction of SARS-CoV-2 receptor binding domain in co... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22738 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
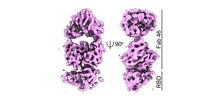
| Title | CryoEM reconstruction of SARS-CoV-2 receptor binding domain in complex with the Fab fragment of neutralizing antibody 46 | ||||||||||||||||||
 Map data Map data | Fab 46 in complex with RBD domain of SARS-CoV-2 Spike protein. | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | ||||||||||||||||||
 Authors Authors | Kucharska I / Tan YZ / Benlekbir S / Rubinstein JL / Julien JP | ||||||||||||||||||
| Funding support |  Canada, 5 items Canada, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers. Authors: Edurne Rujas / Iga Kucharska / Yong Zi Tan / Samir Benlekbir / Hong Cui / Tiantian Zhao / Gregory A Wasney / Patrick Budylowski / Furkan Guvenc / Jocelyn C Newton / Taylor Sicard / Anthony ...Authors: Edurne Rujas / Iga Kucharska / Yong Zi Tan / Samir Benlekbir / Hong Cui / Tiantian Zhao / Gregory A Wasney / Patrick Budylowski / Furkan Guvenc / Jocelyn C Newton / Taylor Sicard / Anthony Semesi / Krithika Muthuraman / Amy Nouanesengsy / Clare Burn Aschner / Katherine Prieto / Stephanie A Bueler / Sawsan Youssef / Sindy Liao-Chan / Jacob Glanville / Natasha Christie-Holmes / Samira Mubareka / Scott D Gray-Owen / John L Rubinstein / Bebhinn Treanor / Jean-Philippe Julien /    Abstract: SARS-CoV-2, the virus responsible for COVID-19, has caused a global pandemic. Antibodies can be powerful biotherapeutics to fight viral infections. Here, we use the human apoferritin protomer as a ...SARS-CoV-2, the virus responsible for COVID-19, has caused a global pandemic. Antibodies can be powerful biotherapeutics to fight viral infections. Here, we use the human apoferritin protomer as a modular subunit to drive oligomerization of antibody fragments and transform antibodies targeting SARS-CoV-2 into exceptionally potent neutralizers. Using this platform, half-maximal inhibitory concentration (IC) values as low as 9 × 10 M are achieved as a result of up to 10,000-fold potency enhancements compared to corresponding IgGs. Combination of three different antibody specificities and the fragment crystallizable (Fc) domain on a single multivalent molecule conferred the ability to overcome viral sequence variability together with outstanding potency and IgG-like bioavailability. The MULTi-specific, multi-Affinity antiBODY (Multabody or MB) platform thus uniquely leverages binding avidity together with multi-specificity to deliver ultrapotent and broad neutralizers against SARS-CoV-2. The modularity of the platform also makes it relevant for rapid evaluation against other infectious diseases of global health importance. Neutralizing antibodies are a promising therapeutic for SARS-CoV-2. #1:  Journal: Biorxiv / Year: 2020 Journal: Biorxiv / Year: 2020Title: Multivalency transforms SARS-CoV-2 antibodies into broad and ultrapotent neutralizers Authors: Rujas E / Kucharska I / Tan YZ / Benlekbir S / Cui H / Zhao T / Wasney G / Budylowski P / Guvenc F / Newton J / Semesi A / Muthuraman K / Nouanesengsy A / Prieto K / Bueler S / Youssef S / ...Authors: Rujas E / Kucharska I / Tan YZ / Benlekbir S / Cui H / Zhao T / Wasney G / Budylowski P / Guvenc F / Newton J / Semesi A / Muthuraman K / Nouanesengsy A / Prieto K / Bueler S / Youssef S / Liao-Chan S / Glanville J / Christie-Holmes N / Mubareka S / Gray-Owen SD / Rubinstein JL / Treanor B / Julien JP | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22738.map.gz emd_22738.map.gz | 28.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22738-v30.xml emd-22738-v30.xml emd-22738.xml emd-22738.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22738.png emd_22738.png | 178.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22738 http://ftp.pdbj.org/pub/emdb/structures/EMD-22738 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22738 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22738 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22738.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22738.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Fab 46 in complex with RBD domain of SARS-CoV-2 Spike protein. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SARS-CoV-2 receptor binding domain in complex with the Fab fragme...
| Entire | Name: SARS-CoV-2 receptor binding domain in complex with the Fab fragment of neutralizing antibody 46 |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 receptor binding domain in complex with the Fab fragme...
| Supramolecule | Name: SARS-CoV-2 receptor binding domain in complex with the Fab fragment of neutralizing antibody 46 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Fab fragment of neutralizing antibody 46
| Supramolecule | Name: Fab fragment of neutralizing antibody 46 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: SARS-CoV-2 receptor binding domain
| Supramolecule | Name: SARS-CoV-2 receptor binding domain / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Fab 46 heavy chain
| Macromolecule | Name: Fab 46 heavy chain / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLLESGGG LVQPGRSLRL SCAASGFTFS SYAMSWVRQA PGKGLEWVST IYSGGSTYY ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGD S RDAFDIWG QGTMVTVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD YF PEPVTVS WNSGALTSGV ...String: EVQLLESGGG LVQPGRSLRL SCAASGFTFS SYAMSWVRQA PGKGLEWVST IYSGGSTYY ADSVKGRFTI SRDNSKNTLY LQMNSLRAED TAVYYCARGD S RDAFDIWG QGTMVTVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD YF PEPVTVS WNSGALTSGV HTFPAVLQSS GLYSLSSVVT VPSSSLGTQT YIC NVNHKP SNTKVDKKVE PKSC |
-Macromolecule #2: Fab 46 light chain
| Macromolecule | Name: Fab 46 light chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ITCRASQSIS SWLAWYQQKP GKAPKLLIYD ASNLETGVP SRFSGSGSGT DFTLTISSLQ PEDFATYYCQ QSYSTPFTFG P GTKVDIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KV DNALQSG NSQESVTEQD ...String: DIQMTQSPSS LSASVGDRVT ITCRASQSIS SWLAWYQQKP GKAPKLLIYD ASNLETGVP SRFSGSGSGT DFTLTISSLQ PEDFATYYCQ QSYSTPFTFG P GTKVDIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KV DNALQSG NSQESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQG LSSPVT KSFNRGEC |
-Macromolecule #3: Receptor Binding Domain of SARS-CoV-2 Spike
| Macromolecule | Name: Receptor Binding Domain of SARS-CoV-2 Spike / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RVQPTESIVR FPNITNLCPF GEVFNATRFA SVYAWNRKRI SNCVADYSVL YNSASFSTFK CYGVSPTKLN DLCFTNVYAD SFVIRGDEVR QIAPGQTGKI ADYNYKLPDD FTGCVIAWNS NNLDSKVGGN YNYLYRLFRK SNLKPFERDI STEIYQAGST PCNGVEGFNC ...String: RVQPTESIVR FPNITNLCPF GEVFNATRFA SVYAWNRKRI SNCVADYSVL YNSASFSTFK CYGVSPTKLN DLCFTNVYAD SFVIRGDEVR QIAPGQTGKI ADYNYKLPDD FTGCVIAWNS NNLDSKVGGN YNYLYRLFRK SNLKPFERDI STEIYQAGST PCNGVEGFNC YFPLQSYGFQ PTNGVGYQPY RVVVLSFELL HAPATVCGPK KSTNLVKNKC VNFHHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Homemade / Material: GOLD / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: glow discharged with PELCO easiGlow (Ted Pella) | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 4722 / Average exposure time: 9.6 sec. / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















