-Search query
-Search result
Showing 1 - 50 of 110 items for (author: ryu & b)
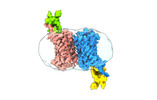
EMDB-36488: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map)

EMDB-37212: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Receptor original map)
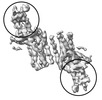
EMDB-37214: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Ligand/CCL7 focused map)

PDB-8jps: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map)

EMDB-37712: 
Small-heat shock protein from Methanocaldococcus jannaschii, Hsp16.5

EMDB-37713: 
Small-heat shock protein from Methanocaldococcus jannaschii, Hsp16.5

PDB-8wp9: 
Small-heat shock protein from Methanocaldococcus jannaschii, Hsp16.5

EMDB-42149: 
S1V2-72 Fab bound to EHA2 from influenza B/Malaysia/2506/2004

PDB-8udg: 
S1V2-72 Fab bound to EHA2 from influenza B/Malaysia/2506/2004

EMDB-33274: 
Human Cx36/GJD2 (N-terminal deletion mutant) gap junction channel in soybean lipids (D6 symmetry)

PDB-7xl8: 
Human Cx36/GJD2 (N-terminal deletion mutant) gap junction channel in soybean lipids (D6 symmetry)

EMDB-36220: 
Cryo-EM structure of Na+,K+-ATPase in the E1.Mg2+ state.

PDB-8jfz: 
Cryo-EM structure of Na+,K+-ATPase in the E1.Mg2+ state.

EMDB-33275: 
Human Cx36/GJD2 (N-terminal deletion mutant) gap junction channel in soybean lipids (C1 symmetry)

EMDB-33326: 
Human Cx36/GJD2 gap junction channel with pore-lining N-terminal helices in the presence of Magnesium

EMDB-33256: 
Human Cx36/GJD2 gap junction channel in detergents

PDB-7xkk: 
Human Cx36/GJD2 gap junction channel in detergents

EMDB-33254: 
Human Cx36/GJD2 (N-terminal deletion BRIL-fused mutant) gap junction channel in soybean lipids (D6 symmetry)

EMDB-33255: 
Human Cx36/GJD2 (N-terminal deletion BRIL-fused mutant) gap junction channel in soybean lipids (C1 symmetry)

EMDB-33270: 
Human Cx36/GJD2 (BRIL-fused mutant) gap junction channel in detergents at 2.2 Angstroms resolution

EMDB-33315: 
Human Cx36/GJD2 gap junction channel with pore-lining N-terminal helices in soybean lipids

EMDB-33327: 
Structurally hetero-junctional human Cx36/GJD2 gap junction channel in soybean lipids (C6 symmetry)

EMDB-33328: 
Structurally hetero-junctional human Cx36/GJD2 gap junction channel in soybean lipids (C1 symmetry)

EMDB-34822: 
Human Cx36/GJD2 (BRIL-fused mutant) gap junction channel in soybean lipids

EMDB-34856: 
Structurally hetero-junctional human Cx36/GJD2 gap junction channel in detergents (C6 symmetry)

EMDB-34857: 
Structurally hetero-junctional human Cx36/GJD2 gap junction channel in detergents (C1 symmetry)

PDB-7xki: 
Human Cx36/GJD2 (N-terminal deletion BRIL-fused mutant) gap junction channel in soybean lipids (D6 symmetry)

PDB-7xkt: 
Human Cx36/GJD2 (BRIL-fused mutant) gap junction channel in detergents at 2.2 Angstroms resolution

PDB-7xnh: 
Human Cx36/GJD2 gap junction channel with pore-lining N-terminal helices in soybean lipids

PDB-7xnv: 
Structurally hetero-junctional human Cx36/GJD2 gap junction channel in soybean lipids (C6 symmetry)

PDB-8hkp: 
Structurally hetero-junctional human Cx36/GJD2 gap junction channel in detergents (C6 symmetry)

EMDB-32892: 
Native Photosystem I of Chlamydomonas reinhardtii

EMDB-32907: 
PSI-LHCI from Chlamydomonas reinhardtii with bound ferredoxin

PDB-7wyi: 
Native Photosystem I of Chlamydomonas reinhardtii

PDB-7wzn: 
PSI-LHCI from Chlamydomonas reinhardtii with bound ferredoxin

EMDB-33293: 
Cryo-EM structure of Cytochrome bo3 from Escherichia coli, apo structure with DMSO

EMDB-33294: 
Cryo-EM structure of Cytochrome bo3 from Escherichia coli, the structure complexed with an allosteric inhibitor N4

PDB-7xmc: 
Cryo-EM structure of Cytochrome bo3 from Escherichia coli, apo structure with DMSO

PDB-7xmd: 
Cryo-EM structure of Cytochrome bo3 from Escherichia coli, the structure complexed with an allosteric inhibitor N4

EMDB-14634: 
Cryo-EM structure of GMPCPP-microtubules in complex with VASH2-SVBP

PDB-7zcw: 
Cryo-EM structure of GMPCPP-microtubules in complex with VASH2-SVBP

EMDB-32097: 
Inward-facing structure of human EAAT2 in the WAY213613-bound state

EMDB-32098: 
Inward-facing structure of human EAAT2 in the substrate-free state

PDB-7vr7: 
Inward-facing structure of human EAAT2 in the WAY213613-bound state

PDB-7vr8: 
Inward-facing structure of human EAAT2 in the substrate-free state

EMDB-33601: 
Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state

EMDB-33602: 
Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state after addition of ATP

PDB-7y45: 
Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state

PDB-7y46: 
Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state after addition of ATP

EMDB-14626: 
Human elongator Elp456 subcomplex
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
