[English] 日本語
 Yorodumi
Yorodumi- EMDB-33270: Human Cx36/GJD2 (BRIL-fused mutant) gap junction channel in deter... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Cx36/GJD2 (BRIL-fused mutant) gap junction channel in detergents at 2.2 Angstroms resolution | |||||||||
 Map data Map data | sharpened_map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | connexin 36 / Cx36 / Gap Junction Channel / BRIL / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationElectric Transmission Across Gap Junctions / connexin complex / Gap junction assembly / gap junction channel activity / neuronal action potential / visual perception / electron transport chain / cell-cell signaling / chemical synaptic transmission / electron transfer activity ...Electric Transmission Across Gap Junctions / connexin complex / Gap junction assembly / gap junction channel activity / neuronal action potential / visual perception / electron transport chain / cell-cell signaling / chemical synaptic transmission / electron transfer activity / periplasmic space / iron ion binding / heme binding / synapse / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.2 Å | |||||||||
 Authors Authors | Cho HJ / Lee SN / Jeong H / Ryu B / Lee HJ / Woo JS / Lee HH | |||||||||
| Funding support |  Korea, Republic Of, 2 items Korea, Republic Of, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel. Authors: Seu-Na Lee / Hwa-Jin Cho / Hyeongseop Jeong / Bumhan Ryu / Hyuk-Joon Lee / Minsoo Kim / Jejoong Yoo / Jae-Sung Woo / Hyung Ho Lee /  Abstract: Connexin 36 (Cx36) is responsible for signal transmission in electrical synapses by forming interneuronal gap junctions. Despite the critical role of Cx36 in normal brain function, the molecular ...Connexin 36 (Cx36) is responsible for signal transmission in electrical synapses by forming interneuronal gap junctions. Despite the critical role of Cx36 in normal brain function, the molecular architecture of the Cx36 gap junction channel (GJC) is unknown. Here, we determine cryo-electron microscopy structures of Cx36 GJC at 2.2-3.6 Å resolutions, revealing a dynamic equilibrium between its closed and open states. In the closed state, channel pores are obstructed by lipids, while N-terminal helices (NTHs) are excluded from the pore. In the open state with pore-lining NTHs, the pore is more acidic than those in Cx26 and Cx46/50 GJCs, explaining its strong cation selectivity. The conformational change during channel opening also includes the α-to-π-helix transition of the first transmembrane helix, which weakens the protomer-protomer interaction. Our structural analyses provide high resolution information on the conformational flexibility of Cx36 GJC and suggest a potential role of lipids in the channel gating. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33270.map.gz emd_33270.map.gz | 566.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33270-v30.xml emd-33270-v30.xml emd-33270.xml emd-33270.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33270.png emd_33270.png | 122.2 KB | ||
| Filedesc metadata |  emd-33270.cif.gz emd-33270.cif.gz | 5.7 KB | ||
| Others |  emd_33270_half_map_1.map.gz emd_33270_half_map_1.map.gz emd_33270_half_map_2.map.gz emd_33270_half_map_2.map.gz | 557.4 MB 557.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33270 http://ftp.pdbj.org/pub/emdb/structures/EMD-33270 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33270 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33270 | HTTPS FTP |
-Related structure data
| Related structure data |  7xktMC  7xkiC  7xkkC  7xl8C  7xnhC  7xnvC  8hkpC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33270.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33270.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened_map | ||||||||||||||||||||||||||||||||||||
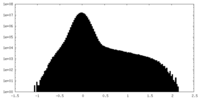
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.675 Å | ||||||||||||||||||||||||||||||||||||
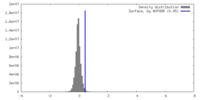
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map A
| File | emd_33270_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_map_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
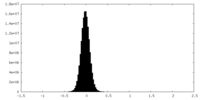
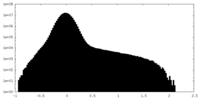
| Density Histograms |
-Half map: half map B
| File | emd_33270_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_map_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
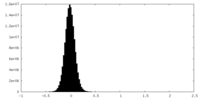
| Density Histograms |
- Sample components
Sample components
-Entire : Cx36-BRIL
| Entire | Name: Cx36-BRIL |
|---|---|
| Components |
|
-Supramolecule #1: Cx36-BRIL
| Supramolecule | Name: Cx36-BRIL / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Gap junction delta-2 protein,Soluble cytochrome b562
| Macromolecule | Name: Gap junction delta-2 protein,Soluble cytochrome b562 / type: protein_or_peptide / ID: 1 Details: The chimeric protein of Cx36 (UNP residues 1-108), BRIL (UNP residues 23-123 with mutation M29W, H124I, R128L), Cx36 (UNP residues 188-321), linker and tags Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.999199 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGEWTILERL LEAAVQQHST MIGRILLTVV VIFRILIVAI VGETVYDDEQ TMFVCNTLQP GCNQACYDRA FPISHIRYWV FQIIMVCTP SLCFITYSVH QSAKQRERRA DLEDNWETLN DNLKVIEKAD NAAQVKDALT KMRAAALDAQ KATPPKLEDK S PDSPEMKD ...String: MGEWTILERL LEAAVQQHST MIGRILLTVV VIFRILIVAI VGETVYDDEQ TMFVCNTLQP GCNQACYDRA FPISHIRYWV FQIIMVCTP SLCFITYSVH QSAKQRERRA DLEDNWETLN DNLKVIEKAD NAAQVKDALT KMRAAALDAQ KATPPKLEDK S PDSPEMKD FRHGFDILVG QIDDALKLAN EGKVKEAQAA AEQLKTTRNA YIQKYLKLRR QEGISRFYII QVVFRNALEI GF LVGQYFL YGFSVPGLYE CNRYPCIKEV ECYVSRPTEK TVFLVFMFAV SGICVVLNLA ELNHLGWRKI KLAVRGAQAK RKS IYEIRN KDLPRVSVPN FGRTQSSDSA YVSRDYKDDD DK UniProtKB: Gap junction delta-2 protein, Soluble cytochrome b562, Gap junction delta-2 protein |
-Macromolecule #2: 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 2 / Number of copies: 72 / Formula: MC3 |
|---|---|
| Molecular weight | Theoretical: 677.933 Da |
| Chemical component information |  ChemComp-MC3: |
-Macromolecule #3: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 3 / Number of copies: 24 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #4: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
| Macromolecule | Name: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE / type: ligand / ID: 4 / Number of copies: 12 / Formula: LHG |
|---|---|
| Molecular weight | Theoretical: 722.97 Da |
| Chemical component information |  ChemComp-LHG: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 444 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 70095 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)