-Search query
-Search result
Showing 1 - 50 of 187 items for (author: loo & ja)

EMDB-16375: 
SARS-CoV2 Omicron BA.1 RBD in complex with CAB-A17 antibody

PDB-8c0y: 
SARS-CoV2 Omicron BA.1 RBD in complex with CAB-A17 antibody

EMDB-42970: 
Model and map from local refinement of a CAB-A17 - Omicron Ba.1 spike complex

PDB-8v4f: 
Model and map from local refinement of a CAB-A17 - Omicron Ba.1 spike complex
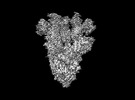
EMDB-28198: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with LLNL-199

EMDB-28199: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with 2130-1-0114-112

PDB-8ekd: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with 2130-1-0114-112

EMDB-17659: 
ACAD9-WT in complex with ECSIT-CTER

EMDB-17660: 
Cryo-EM structure of human ACAD9-S191A

EMDB-17661: 
ACAD9 homodimer WT

PDB-8phe: 
ACAD9-WT in complex with ECSIT-CTER

PDB-8phf: 
Cryo-EM structure of human ACAD9-S191A

EMDB-41825: 
Prefusion-stabilized Langya virus F protein, variant G99C/I109C

EMDB-42940: 
Dimer of Hendra virus prefusion F trimers

EMDB-42942: 
Dimer of Langya virus prefusion F trimers

PDB-8u1r: 
Prefusion-stabilized Langya virus F protein, variant G99C/I109C

EMDB-26583: 
Cryo-EM structure of Antibody 12-16 in complex with prefusion SARS-CoV-2 Spike glycoprotein

PDB-7ukl: 
Cryo-EM structure of Antibody 12-16 in complex with prefusion SARS-CoV-2 Spike glycoprotein

EMDB-40789: 
BAP1/ASXL1 bound to the H2AK119Ub Nucleosome

EMDB-40790: 
Map focused on acidic patch BAP1/ASXL1 bound to the H2AK119Ub Nucleosome

EMDB-40791: 
Overall map of BAP1/ASXL1 bound to the H2AK119Ub Nucleosome

PDB-8svf: 
BAP1/ASXL1 bound to the H2AK119Ub Nucleosome

EMDB-27566: 
Prefusion-stabilized Nipah virus fusion protein

EMDB-27577: 
Prefusion-stabilized Hendra virus fusion protein

EMDB-27590: 
Prefusion-stabilized Nipah virus fusion protein, dimer of trimers

PDB-8dng: 
Prefusion-stabilized Nipah virus fusion protein

PDB-8dnr: 
Prefusion-stabilized Hendra virus fusion protein

PDB-8do4: 
Prefusion-stabilized Nipah virus fusion protein, dimer of trimers

EMDB-26652: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 4H3

EMDB-26658: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 2D3

EMDB-26659: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1H8

EMDB-26660: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1H1

EMDB-26662: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 2B12

EMDB-26668: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1A9

PDB-7uop: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 4H3

PDB-7up9: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 2D3

PDB-7upa: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1H8

PDB-7upb: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1H1

PDB-7upd: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 2B12

PDB-7upk: 
Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1A9

EMDB-29052: 
SARS-CoV-2 Spike Hexapro - C68.59 Fab (Class 3 - disordered)

EMDB-29053: 
SARS-CoV-2 Spike Hexapro - C59.68 Fab (Class 1 - No Fab bound)

EMDB-29054: 
SARS-CoV-2 Spike Hexapro - C68.59 Fab (Class 2 - Fab bound)
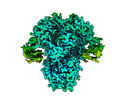
EMDB-26727: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation
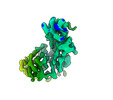
EMDB-26729: 
CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0)
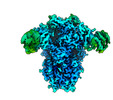
EMDB-26730: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation
EMDB-26731: 
CCoV-HuPn-2018 S in the swung out conformation (local refinement of domain 0)

PDB-7us6: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation

PDB-7us9: 
CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0)

PDB-7usa: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
