-Search query
-Search result
Showing 1 - 50 of 61 items for (author: loeff & l)

EMDB-50090: 
Vibrio cholerae DdmD apo complex

PDB-9ezx: 
Vibrio cholerae DdmD apo complex

EMDB-18539: 
Structure of the human 80S ribosome at 1.9 A resolution - the molecular role of chemical modifications and ions in RNA
EMDB-18812: 
The structure of the human 80S ribosome at 1.9 angstrom resolution reveals the molecular role of chemical modifications and ions in RNA - Focused refinement of the of the 60S subunit
EMDB-18813: 
The structure of the human 80S ribosome at 1.9 angstrom resolution reveals the molecular role of chemical modifications and ions in RNA - Focused refinement of the of the 40S subunit body
EMDB-18814: 
The structure of the human 80S ribosome at 1.9 angstrom resolution reveals the molecular role of chemical modifications and ions in RNA - Focused refinement of the of the 40S subunit head
EMDB-18815: 
Structure of the human 80S ribosome at 1.9 A resolution - the molecular role of chemical modifications and ions in RNA - Global human 80S ribosome refinement before focused refinements.

PDB-8qoi: 
Structure of the human 80S ribosome at 1.9 A resolution - the molecular role of chemical modifications and ions in RNA

EMDB-14493: 
SpCas9 bound to 6 nucleotide complementary DNA substrate

EMDB-14494: 
SpCas9 bound to 8-nucleotide complementary DNA substrate
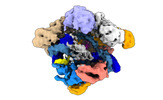
EMDB-14496: 
SpCas9 bound to 12-nucleotide complementary DNA substrate
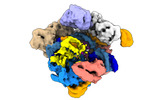
EMDB-14497: 
SpCas9 bound to 14-nucleotide complementary DNA substrate

EMDB-14498: 
SpCas9 bound to 16-nucleotide complementary DNA substrate
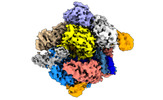
EMDB-14499: 
SpCas9 bound to 18-nucleotide complementary DNA substrate in the catalytic state
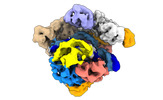
EMDB-14500: 
SpCas9 bound to 10-nucleotide complementary DNA substrate

EMDB-14501: 
SpCas9 bound to 18-nucleotide complementary DNA substrate in the checkpoint state

PDB-7z4c: 
SpCas9 bound to 6 nucleotide complementary DNA substrate

PDB-7z4e: 
SpCas9 bound to 8-nucleotide complementary DNA substrate

PDB-7z4g: 
SpCas9 bound to 12-nucleotide complementary DNA substrate

PDB-7z4h: 
SpCas9 bound to 14-nucleotide complementary DNA substrate

PDB-7z4i: 
SpCas9 bound to 16-nucleotide complementary DNA substrate

PDB-7z4j: 
SpCas9 bound to 18-nucleotide complementary DNA substrate in the catalytic state

PDB-7z4k: 
SpCas9 bound to 10-nucleotide complementary DNA substrate

PDB-7z4l: 
SpCas9 bound to 18-nucleotide complementary DNA substrate in the checkpoint state

EMDB-13976: 
Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (stigmatellin and azide bound)

EMDB-13977: 
Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (as isolated)

PDB-7qhm: 
Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (stigmatellin and azide bound)

PDB-7qho: 
Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (as isolated)

EMDB-10959: 
Cryo-EM structure of the ARP2/3 1B5CL isoform complex.

EMDB-10960: 
Cryo-EM structure of the ARP2/3 1A5C isoform complex.

PDB-6yw6: 
Cryo-EM structure of the ARP2/3 1B5CL isoform complex.

PDB-6yw7: 
Cryo-EM structure of the ARP2/3 1A5C isoform complex.

EMDB-10668: 
Structure of human ribosome in classical-PRE state

EMDB-10674: 
Cryo-EM map of human ribosome in POST state

EMDB-10690: 
Cryo-EM map of human ribosome in hybrid-PRE state

PDB-6y0g: 
Structure of human ribosome in classical-PRE state

PDB-6y2l: 
Structure of human ribosome in POST state

PDB-6y57: 
Structure of human ribosome in hybrid-PRE state

PDB-6s8m: 
S. pombe microtubule decorated with Cut7 motor domain in the AMPPNP state
EMDB-0324: 
Negative-stain map of CPFcore

EMDB-0325: 
Cryo-EM map of the Ysh1-Mpe1 nuclease complex

EMDB-3530: 
Ustilago maydis kinesin-5 motor domain with N-terminal extension in the AMPPNP state bound to microtubules

PDB-5mm7: 
Ustilago maydis kinesin-5 motor domain with N-terminal extension in the AMPPNP state bound to microtubules

EMDB-3527: 
S. pombe microtubule decorated with Cut7 motor domain in the AMPPNP state

EMDB-3529: 
Ustilago maydis kinesin-5 motor domain in the AMPPNP state bound to microtubules

PDB-5mm4: 
Ustilago maydis kinesin-5 motor domain in the AMPPNP state bound to microtubules

EMDB-4260: 
Volta phase plate data collection facilitates image processing and cryo-EM structure determination

EMDB-4261: 
Volta phase plate data collection facilitates image processing and cryo-EM structure determination

EMDB-4262: 
Volta phase plate data collection facilitates image processing and cryo-EM structure determination

EMDB-3522: 
S. pombe microtubule copolymerized with GTP and Mal3-143
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
