+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0324 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative-stain map of CPFcore | |||||||||
 Map data Map data | Negative-stain map of CPFcore | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
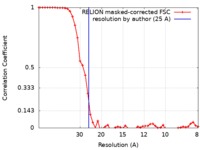
| Method | single particle reconstruction / negative staining / Resolution: 25.0 Å | |||||||||
 Authors Authors | Hill CH / Boreikaite V / Kumar A / Casanal A / Kubik P / Degliesposti G / Maslen S / Mariani A / von Loeffelholz O / Girbig M ...Hill CH / Boreikaite V / Kumar A / Casanal A / Kubik P / Degliesposti G / Maslen S / Mariani A / von Loeffelholz O / Girbig M / Skehel M / Passmore LA | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2019 Journal: Mol Cell / Year: 2019Title: Activation of the Endonuclease that Defines mRNA 3' Ends Requires Incorporation into an 8-Subunit Core Cleavage and Polyadenylation Factor Complex. Authors: Chris H Hill / Vytautė Boreikaitė / Ananthanarayanan Kumar / Ana Casañal / Peter Kubík / Gianluca Degliesposti / Sarah Maslen / Angelica Mariani / Ottilie von Loeffelholz / Mathias ...Authors: Chris H Hill / Vytautė Boreikaitė / Ananthanarayanan Kumar / Ana Casañal / Peter Kubík / Gianluca Degliesposti / Sarah Maslen / Angelica Mariani / Ottilie von Loeffelholz / Mathias Girbig / Mark Skehel / Lori A Passmore /   Abstract: Cleavage and polyadenylation factor (CPF/CPSF) is a multi-protein complex essential for formation of eukaryotic mRNA 3' ends. CPF cleaves pre-mRNAs at a specific site and adds a poly(A) tail. The ...Cleavage and polyadenylation factor (CPF/CPSF) is a multi-protein complex essential for formation of eukaryotic mRNA 3' ends. CPF cleaves pre-mRNAs at a specific site and adds a poly(A) tail. The cleavage reaction defines the 3' end of the mature mRNA, and thus the activity of the endonuclease is highly regulated. Here, we show that reconstitution of specific pre-mRNA cleavage with recombinant yeast proteins requires incorporation of the Ysh1 endonuclease into an eight-subunit "CPF" complex. Cleavage also requires the accessory cleavage factors IA and IB, which bind substrate pre-mRNAs and CPF, likely facilitating assembly of an active complex. Using X-ray crystallography, electron microscopy, and mass spectrometry, we determine the structure of Ysh1 bound to Mpe1 and the arrangement of subunits within CPF. Together, our data suggest that the active mRNA 3' end processing machinery is a dynamic assembly that is licensed to cleave only when all protein factors come together at the polyadenylation site. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0324.map.gz emd_0324.map.gz | 1.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0324-v30.xml emd-0324-v30.xml emd-0324.xml emd-0324.xml | 24.3 KB 24.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0324_fsc.xml emd_0324_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_0324.png emd_0324.png | 47.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0324 http://ftp.pdbj.org/pub/emdb/structures/EMD-0324 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0324 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0324 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0324.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0324.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative-stain map of CPFcore | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.98 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CPFcore: a complex of Cft1-Pfs2-Yth1-Fip1-Pap1-Cft2-Ysh1-Mpe1
| Entire | Name: CPFcore: a complex of Cft1-Pfs2-Yth1-Fip1-Pap1-Cft2-Ysh1-Mpe1 |
|---|---|
| Components |
|
-Supramolecule #1: CPFcore: a complex of Cft1-Pfs2-Yth1-Fip1-Pap1-Cft2-Ysh1-Mpe1
| Supramolecule | Name: CPFcore: a complex of Cft1-Pfs2-Yth1-Fip1-Pap1-Cft2-Ysh1-Mpe1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
-Macromolecule #1: Cft1
| Macromolecule | Name: Cft1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MNVYDDVLDA TVVSHSLATH FTTSDYEELL VVRTNILSVY RPTRDGKLYL TDEFKFHGLI TDIGLIPQK DSPLSCLLLC TGVAKISILK FNTLTNSIDT LSLHYYEGKF KGKSLVELAK I STLRMDPG SSCALLFNND IIAFLPFHVN KNDDDEEEED EDENIDDSEL ...String: MNVYDDVLDA TVVSHSLATH FTTSDYEELL VVRTNILSVY RPTRDGKLYL TDEFKFHGLI TDIGLIPQK DSPLSCLLLC TGVAKISILK FNTLTNSIDT LSLHYYEGKF KGKSLVELAK I STLRMDPG SSCALLFNND IIAFLPFHVN KNDDDEEEED EDENIDDSEL IHSMNQKSQG TN TFNKRKR TKLGDKFTAP SVVLVASELY EGAKNIIDIQ FLKNFTKPTI ALLYQPKLVW AGN TTISKL PTQYVILTLN IQPAESATKI ESTTIAFVKE LPWDLHTIVP VSNGAIIVGT NELA FLDNT GVLQSTVLLN SFADKELQKT KIINNSSLEI MFREKNTTSI WIPSSKSKNG GSNND ETLL LMDLKSNIYY IQMEAEGRLL IKFDIFKLPI VNDLLKENSN PKCITRLNAT NSNKNM DLF IGFGSGNALV LRLNNLKSTI ETREAHNPSS GTNSLMDIND DDDEEMDDLY ADEAPEN GL TTNDSKGTVE TVQPFDIELL SSLRNVGPIT SLTVGKVSSI DDVVKGLPNP NKNEYSLV A TSGNGSGSHL TVIQTSVQPE IELALKFISI TQIWNLKIKG RDRYLITTDS TKSRSDIYE SDNNFKLHKG GRLRRDATTV YISMFGEEKR IIQVTTNHLY LYDTHFRRLT TIKFDYEVIH VSVMDPYIL VTVSRGDIKI FELEEKNKRK LLKVDLPEIL NEMVITSGLI LKSNMCNEFL I GLSKSQEE QLLFTFVTAD NQIIFFTKDH NDRIFQLNGV DQLNESLYIS TYQLGDEIVP DP SIKQVMI NKLGHDNKEE YLTILTFGGE IYQYRKLPQR RSRFYRNVTR NDLAITGAPD NAY AKGVSS IERIMHYFPD YNGYSVIFVT GSVPYILIKE DDSTPKIFKF GNIPLVSVTP WSER SVMCV DDIKNARVYT LTTDNMYYGN KLPLKQIKIS NVLDDYKTLQ KLVYHERAQL FLVSY CKRV PYEALGEDGE KVIGYDENVP HAEGFQSGIL LINPKSWKVI DKIDFPKNSV VNEMRS SMI QINSKTKRKR EYIIAGVANA TTEDTPPTGA FHIYDVIEVV PEPGKPDTNY KLKEIFQ EE VSGTVSTVCE VSGRFMISQS QKVLVRDIQE DNSVIPVAFL DIPVFVTDSK SFGNLLII G DAMQGFQFIG FDAEPYRMIS LGRSMSKFQT MSLEFLVNGG DMYFAATDAD RNVHVLKYA PDEPNSLSGQ RLVHCSSFTL HSTNSCMMLL PRNEEFGSPQ VPSFQNVGGQ VDGSVFKIVP LSEEKYRRL YVIQQQIIDR ELQLGGLNPR MERLANDFYQ MGHSMRPMLD FNVIRRFCGL A IDRRKSIA QKAGRHAHFE AWRDIINIEF SMRSLCQGK |
-Macromolecule #2: Pfs2
| Macromolecule | Name: Pfs2 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MDGHNQNQYQ NQNQIQQSQQ PPLKKYVTQR RSVDVSSPYI NLYYNRRHGL PNLVVEPETS YTIDIMPPN AYRGRDRVIN LPSKFTHLSS NKVKHVIPAI QWTPEGRRLV VATYSGEFSL W NASSFTFE TLMQAHDSAV TTMKYSHDSD WMISGDADGM IKIWQPNFSM ...String: MDGHNQNQYQ NQNQIQQSQQ PPLKKYVTQR RSVDVSSPYI NLYYNRRHGL PNLVVEPETS YTIDIMPPN AYRGRDRVIN LPSKFTHLSS NKVKHVIPAI QWTPEGRRLV VATYSGEFSL W NASSFTFE TLMQAHDSAV TTMKYSHDSD WMISGDADGM IKIWQPNFSM VKEIDAAHTE SI RDMAFSS NDSKFVTCSD DNILKIWNFS NGKQERVLSG HHWDVKSCDW HPEMGLIASA SKD NLVKLW DPRSGNCISS ILKFKHTVLK TRFQPTKGNL LMAISKDKSC RVFDIRYSMK ELMC VRDET DYMTLEWHPI NESMFTLACY DGSLKHFDLL QNLNEPILTI PYAHDKCITS LSYNP VGHI FATAAKDRTI RFWTRARPID PNAYDDPTYN NKKINGWFFG INNDINAVRE KSEFGA APP PPATLEPHAL PNMNGFINKK PRQEIPGIDS NIKSSTLPGL SI |
-Macromolecule #3: Yth1
| Macromolecule | Name: Yth1 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MSLIHPDTAK YPFKFEPFLR QEYSFSLDPD RPICEFYNSR EGPKSCPRGP LCPKKHVLPI FQNKIVCRH WLRGLCKKND QCEYLHEYNL RKMPECVFFS KNGYCTQSPD CQYLHIDPAS K IPKCENYE MGFCPLGSSC PRRHIKKVFC QRYMTGFCPL GKDECDMEHP ...String: MSLIHPDTAK YPFKFEPFLR QEYSFSLDPD RPICEFYNSR EGPKSCPRGP LCPKKHVLPI FQNKIVCRH WLRGLCKKND QCEYLHEYNL RKMPECVFFS KNGYCTQSPD CQYLHIDPAS K IPKCENYE MGFCPLGSSC PRRHIKKVFC QRYMTGFCPL GKDECDMEHP QFIIPDEGSK LR IKRDDEI NTRKMDEEKE RRLNAIINGE V |
-Macromolecule #4: Fip1
| Macromolecule | Name: Fip1 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MSSSEDEDDK FLYGSDSELA LPSSKRSRDD EADAGASSNP DIVKRQKFDS PVEETPATAR DDRSDEDIY SDSSDDDSDS DLEVIISLGP DPTRLDAKLL DSYSTAATSS SKDVISVATD V SNTITKTS DERLITEGEA NQGVTATTVK ATESDGNVPK AMTGSIDLDK ...String: MSSSEDEDDK FLYGSDSELA LPSSKRSRDD EADAGASSNP DIVKRQKFDS PVEETPATAR DDRSDEDIY SDSSDDDSDS DLEVIISLGP DPTRLDAKLL DSYSTAATSS SKDVISVATD V SNTITKTS DERLITEGEA NQGVTATTVK ATESDGNVPK AMTGSIDLDK EGIFDSVGIT TI DPEVLKE KPWRQPGANL SDYFNYGFNE FTWMEYLHRQ EKLQQDYNPR RILMGLLSLQ QQG KLNSAN DTDSNLGNII DNNNNVNNAN MSNLNSNMGN SMSGTPNPPA PPMHPSFPPL PMFG SFPPF PMPGMMPPMN QQPNQNQNQN SK |
-Macromolecule #5: Pap1
| Macromolecule | Name: Pap1 / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MSSQKVFGIT GPVSTVGATA AENKLNDSLI QELKKEGSFE TEQETANRVQ VLKILQELAQ RFVYEVSKK KNMSDGMARD AGGKIFTYGS YRLGVHGPGS DIDTLVVVPK HVTREDFFTV F DSLLRERK ELDEIAPVPD AFVPIIKIKF SGISIDLICA RLDQPQVPLS ...String: MSSQKVFGIT GPVSTVGATA AENKLNDSLI QELKKEGSFE TEQETANRVQ VLKILQELAQ RFVYEVSKK KNMSDGMARD AGGKIFTYGS YRLGVHGPGS DIDTLVVVPK HVTREDFFTV F DSLLRERK ELDEIAPVPD AFVPIIKIKF SGISIDLICA RLDQPQVPLS LTLSDKNLLR NL DEKDLRA LNGTRVTDEI LELVPKPNVF RIALRAIKLW AQRRAVYANI FGFPGGVAWA MLV ARICQL YPNACSAVIL NRFFIILSEW NWPQPVILKP IEDGPLQVRV WNPKIYAQDR SHRM PVITP AYPSMCATHN ITESTKKVIL QEFVRGVQIT NDIFSNKKSW ANLFEKNDFF FRYKF YLEI TAYTRGSDEQ HLKWSGLVES KVRLLVMKLE VLAGIKIAHP FTKPFESSYC CPTEDD YEM IQDKYGSHKT ETALNALKLV TDENKEEESI KDAPKAYLST MYIGLDFNIE NKKEKVD IH IPCTEFVNLC RSFNEDYGDH KVFNLALRFV KGYDLPDEVF DENEKRPSKK SKRKNLDA R HETVKRSKSD AASGDNINGT TAAVDVN |
-Macromolecule #6: Cft2
| Macromolecule | Name: Cft2 / type: protein_or_peptide / ID: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MTYKYNCCDD GSGTTVGSVV RFDNVTLLID PGWNPSKVSY EQCIKYWEKV IPEIDVIILS QPTIECLGA HSLLYYNFTS HFISRIQVYA TLPVINLGRV STIDSYASAG VIGPYDTNKL D LEDIEISF DHIVPLKYSQ LVDLRSRYDG LTLLAYNAGV CPGGSIWCIS ...String: MTYKYNCCDD GSGTTVGSVV RFDNVTLLID PGWNPSKVSY EQCIKYWEKV IPEIDVIILS QPTIECLGA HSLLYYNFTS HFISRIQVYA TLPVINLGRV STIDSYASAG VIGPYDTNKL D LEDIEISF DHIVPLKYSQ LVDLRSRYDG LTLLAYNAGV CPGGSIWCIS TYSEKLVYAK RW NHTRDNI LNAASILDAT GKPLSTLMRP SAIITTLDRF GSSQPFKKRS KIFKDTLKKG LSS DGSVII PVDMSGKFLD LFTQVHELLF ESTKINAHTQ VPVLILSYAR GRTLTYAKSM LEWL SPSLL KTWENRNNTS PFEIGSRIKI IAPNELSKYP GSKICFVSEV GALINEVIIK VGNSE KTTL ILTKPSFECA SSLDKILEIV EQDERNWKTF PEDGKSFLCD NYISIDTIKE EPLSKE ETE AFKVQLKEKK RDRNKKILLV KRESKKLANG NAIIDDTNGE RAMRNQDILV ENVNGVP PI DHIMGGDEDD DEEEENDNLL NLLKDNSEKS AAKKNTEVPV DIIIQPSAAS KHKMFPFN P AKIKKDDYGT VVDFTMFLPD DSDNVNQNSR KRPLKDGAKT TSPVNEEDNK NEEEDGYNM SDPISKRSKH RASRYSGFSG TGEAENFDNL DYLKIDKTLS KRTISTVNVQ LKCSVVILNL QSLVDQRSA SIIWPSLKSR KIVLSAPKQI QNEEITAKLI KKNIEVVNMP LNKIVEFSTT I KTLDISID SNLDNLLKWQ RISDSYTVAT VVGRLVKESL PQVNNHQKTA SRSKLVLKPL HG SSRSHKT GALSIGDVRL AQLKKLLTEK NYIAEFKGEG TLVINEKVAV RKINDAETII DGT PSELFD TVKKLVTDML AKI |
-Macromolecule #7: Ysh1
| Macromolecule | Name: Ysh1 / type: protein_or_peptide / ID: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MERTNTTTFK FFSLGGSNEV GRSCHILQYK GKTVMLDAGI HPAYQGLASL PFYDEFDLSK VDILLISHF HLDHAASLPY VMQRTNFQGR VFMTHPTKAI YRWLLRDFVR VTSIGSSSSS M GTKDEGLF SDEDLVDSFD KIETVDYHST VDVNGIKFTA FHAGHVLGAA ...String: MERTNTTTFK FFSLGGSNEV GRSCHILQYK GKTVMLDAGI HPAYQGLASL PFYDEFDLSK VDILLISHF HLDHAASLPY VMQRTNFQGR VFMTHPTKAI YRWLLRDFVR VTSIGSSSSS M GTKDEGLF SDEDLVDSFD KIETVDYHST VDVNGIKFTA FHAGHVLGAA MFQIEIAGLR VL FTGDYSR EVDRHLNSAE VPPLSSNVLI VESTFGTATH EPRLNRERKL TQLIHSTVMR GGR VLLPVF ALGRAQEIML ILDEYWSQHA DELGGGQVPI FYASNLAKKC MSVFQTYVNM MNDD IRKKF RDSQTNPFIF KNISYLRNLE DFQDFGPSVM LASPGMLQSG LSRDLLERWC PEDKN LVLI TGYSIEGTMA KFIMLEPDTI PSINNPEITI PRRCQVEEIS FAAHVDFQEN LEFIEK ISA PNIILVHGEA NPMGRLKSAL LSNFASLKGT DNEVHVFNPR NCVEVDLEFQ GVKVAKA VG NIVNEIYKEE NVEIKEEIAA KIEPIKEENE DNLDSQAEKG LVDEEEHKDI VVSGILVS D DKNFELDFLS LSDLREHHPD LSTTILRERQ SVRVNCKKEL IYWHILQMFG EAEVLQDDD RVTNQEPKVK EESKDNLTNT GKLILQIMGD IKLTIVNTLA VVEWTQDLMN DTVADSIIAI LMNVDSAPA SVKLSSHSCD DHDHNNVQSN AQGKIDEVER VKQISRLFKE QFGDCFTLFL N KDEYASNK EETITGVVTI GKSTAKIDFN NMKILECNSN PLKGRVESLL NIGGNLVTPL C |
-Macromolecule #8: Mpe1
| Macromolecule | Name: Mpe1 / type: protein_or_peptide / ID: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MSSTIFYRFK SQRNTSRILF DGTGLTVFDL KREIIQENKL GDGTDFQLKI YNPDTEEEYD DDAFVIPRS TSVIVKRSPA IKSFSVHSRL KGNVGAAALG NATRYVTGRP RVLQKRQHTA T TTANVSGT TEEERIASMF ATQENQWEQT QEEMSAATPV FFKSQTNKNS ...String: MSSTIFYRFK SQRNTSRILF DGTGLTVFDL KREIIQENKL GDGTDFQLKI YNPDTEEEYD DDAFVIPRS TSVIVKRSPA IKSFSVHSRL KGNVGAAALG NATRYVTGRP RVLQKRQHTA T TTANVSGT TEEERIASMF ATQENQWEQT QEEMSAATPV FFKSQTNKNS AQENEGPPPP GY MCYRCGG RDHWIKNCPT NSDPNFEGKR IRRTTGIPKK FLKSIEIDPE TMTPEEMAQR KIM ITDEGK FVVQVEDKQS WEDYQRKREN RQIDGDETIW RKGHFKDLPD DLKCPLTGGL LRQP VKTSK CCNIDFSKEA LENALVESDF VCPNCETRDI LLDSLVPDQD KEKEVETFLK KQEEL HGSS KDGNQPETKK MKLMDPTGTA GLNNNTSLPT SVNNGGTPVP PVPLPFGIPP FPMFPM PFM PPTATITNPH QADASPKK |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 Component:
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Acetate Details: Three microliters of sample was applied to the support and allowed to adsorb for 60 s before wicking away with filter paper. Grids were then applied sequentially to two 30 ul drops of 2% w/v ...Details: Three microliters of sample was applied to the support and allowed to adsorb for 60 s before wicking away with filter paper. Grids were then applied sequentially to two 30 ul drops of 2% w/v uranyl acetate, first to wash (quick) and then to stain (30 s). Excess stain was then wicked away with filter paper until dry. | ||||||||||
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 4.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR Details: Grids were CF400-CU-UL from Electron Microscopy Sciences. Thin layer continuous carbon over 400-mesh copper. Manually inspected before use (with a light microscope) | ||||||||||
| Details | 250 nM complex was used |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 1000 (2k x 2k) / Average exposure time: 2.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: -0.6 µm / Nominal defocus min: -0.6 µm / Nominal magnification: 26000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)