+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10960 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
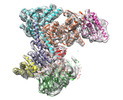
| Title | Cryo-EM structure of the ARP2/3 1A5C isoform complex. | |||||||||
 Map data Map data | Cryo-EM structure of the human Arp2/3 1A5C isoform complex. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cytoskeleton / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmeiotic chromosome movement towards spindle pole / cytosolic transport / growth cone leading edge / spindle localization / meiotic cytokinesis / actin polymerization-dependent cell motility / muscle cell projection membrane / asymmetric cell division / Arp2/3 protein complex / actin nucleation ...meiotic chromosome movement towards spindle pole / cytosolic transport / growth cone leading edge / spindle localization / meiotic cytokinesis / actin polymerization-dependent cell motility / muscle cell projection membrane / asymmetric cell division / Arp2/3 protein complex / actin nucleation / Arp2/3 complex-mediated actin nucleation / actin cap / regulation of actin filament polymerization / filamentous actin / establishment or maintenance of cell polarity / positive regulation of actin filament polymerization / brush border / cilium assembly / RHO GTPases Activate WASPs and WAVEs / regulation of synaptic vesicle endocytosis / positive regulation of double-strand break repair via homologous recombination / positive regulation of lamellipodium assembly / actin filament polymerization / EPHB-mediated forward signaling / positive regulation of substrate adhesion-dependent cell spreading / cell projection / FCGR3A-mediated phagocytosis / cellular response to nerve growth factor stimulus / Regulation of actin dynamics for phagocytic cup formation / structural constituent of cytoskeleton / cellular response to type II interferon / azurophil granule lumen / cell-cell junction / actin filament binding / synaptic vesicle membrane / cell migration / actin cytoskeleton / lamellipodium / Clathrin-mediated endocytosis / site of double-strand break / actin binding / actin cytoskeleton organization / cell cortex / secretory granule lumen / ficolin-1-rich granule lumen / endosome / neuron projection / postsynapse / focal adhesion / Neutrophil degranulation / glutamatergic synapse / enzyme binding / positive regulation of transcription by RNA polymerase II / extracellular exosome / extracellular region / nucleoplasm / ATP binding / nucleus / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | von Loeffelholz O / Moores C | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Biol Open / Year: 2020 Journal: Biol Open / Year: 2020Title: Cryo-EM of human Arp2/3 complexes provides structural insights into actin nucleation modulation by ARPC5 isoforms. Authors: Ottilie von Loeffelholz / Andrew Purkiss / Luyan Cao / Svend Kjaer / Naoko Kogata / Guillaume Romet-Lemonne / Michael Way / Carolyn A Moores /   Abstract: The Arp2/3 complex regulates many cellular processes by stimulating formation of branched actin filament networks. Because three of its seven subunits exist as two different isoforms, mammals produce ...The Arp2/3 complex regulates many cellular processes by stimulating formation of branched actin filament networks. Because three of its seven subunits exist as two different isoforms, mammals produce a family of Arp2/3 complexes with different properties that may be suited to different physiological contexts. To shed light on how isoform diversification affects Arp2/3 function, we determined a 4.2 Å resolution cryo-EM structure of the most active human Arp2/3 complex containing ARPC1B and ARPC5L, and compared it with the structure of the least active ARPC1A-ARPC5-containing complex. The architecture of each isoform-specific Arp2/3 complex is the same. Strikingly, however, the N-terminal half of ARPC5L is partially disordered compared to ARPC5, suggesting that this region of ARPC5/ARPC5L is an important determinant of complex activity. Confirming this idea, the nucleation activity of Arp2/3 complexes containing hybrid ARPC5/ARPC5L subunits is higher when the ARPC5L N-terminus is present, thereby providing insight into activity differences between the different Arp2/3 complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10960.map.gz emd_10960.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10960-v30.xml emd-10960-v30.xml emd-10960.xml emd-10960.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10960.png emd_10960.png | 195.2 KB | ||
| Filedesc metadata |  emd-10960.cif.gz emd-10960.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10960 http://ftp.pdbj.org/pub/emdb/structures/EMD-10960 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10960 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10960 | HTTPS FTP |
-Related structure data
| Related structure data |  6yw7MC  6yw6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10960.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10960.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the human Arp2/3 1A5C isoform complex. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human ARP2/3 1A5C isoform complex
| Entire | Name: human ARP2/3 1A5C isoform complex |
|---|---|
| Components |
|
-Supramolecule #1: human ARP2/3 1A5C isoform complex
| Supramolecule | Name: human ARP2/3 1A5C isoform complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Actin-related protein 3
| Macromolecule | Name: Actin-related protein 3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 47.428031 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGRLPACVV DCGTGYTKLG YAGNTEPQFI IPSCIAIKES AKVGDQAQRR VMKGVDDLDF FIGDEAIEKP TYATKWPIRH GIVEDWDLM ERFMEQVIFK YLRAEPEDHY FLLTEPPLNT PENREYTAEI MFESFNVPGL YIAVQAVLAL AASWTSRQVG E RTLTGTVI ...String: MAGRLPACVV DCGTGYTKLG YAGNTEPQFI IPSCIAIKES AKVGDQAQRR VMKGVDDLDF FIGDEAIEKP TYATKWPIRH GIVEDWDLM ERFMEQVIFK YLRAEPEDHY FLLTEPPLNT PENREYTAEI MFESFNVPGL YIAVQAVLAL AASWTSRQVG E RTLTGTVI DSGDGVTHVI PVAEGYVIGS CIKHIPIAGR DITYFIQQLL RDREVGIPPE QSLETAKAVK ERYSYVCPDL VK EFNKYDT DGSKWIKQYT GINAISKKEF SIDVGYERFL GPEIFFHPEF ANPDFTQPIS EVVDEVIQNC PIDVRRPLYK NIV LSGGST MFRDFGRRLQ RDLKRTVDAR LKLSEELSGG RLKPKPIDVQ VITHHMQRYA VWFGGSMLAS TPEFYQVCHT KKDY EEIGP SICRHNPVFG VMS UniProtKB: Actin-related protein 3 |
-Macromolecule #2: Actin-related protein 2/3 complex subunit 2
| Macromolecule | Name: Actin-related protein 2/3 complex subunit 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.386043 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MILLEVNNRI IEETLALKFE NAAAGNKPEA VEVTFADFDG VLYHISNPNG DKTKVMVSIS LKFYKELQAH GADELLKRVY GSFLVNPES GYNVSLLYDL ENLPASKDSI VHQAGMLKRN CFASVFEKYF QFQEEGKEGE NRAVIHYRDD ETMYVESKKD R VTVVFSTV ...String: MILLEVNNRI IEETLALKFE NAAAGNKPEA VEVTFADFDG VLYHISNPNG DKTKVMVSIS LKFYKELQAH GADELLKRVY GSFLVNPES GYNVSLLYDL ENLPASKDSI VHQAGMLKRN CFASVFEKYF QFQEEGKEGE NRAVIHYRDD ETMYVESKKD R VTVVFSTV FKDDDDVVIG KVFMQEFKEG RRASHTAPQV LFSHREPPLE LKDTDAAVGD NIGYITFVLF PRHTNASARD NT INLIHTF RDYLHYHIKC SKAYIHTRMR AKTSDFLKVL NRARPDAEKK EMKTITGKTF SSR UniProtKB: Actin-related protein 2/3 complex subunit 2 |
-Macromolecule #3: Actin-related protein 2/3 complex subunit 3
| Macromolecule | Name: Actin-related protein 2/3 complex subunit 3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 20.572666 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPAYHSSLMD PDTKLIGNMA LLPIRSQFKG PAPRETKDTD IVDEAIYYFK ANVFFKNYEI KNEADRTLIY ITLYISECLK KLQKCNSKS QGEKEMYTLG ITNFPIPGEP GFPLNAIYAK PANKQEDEVM RAYLQQLRQE TGLRLCEKVF DPQNDKPSKW W TCFVKRQF MNKSLSGPGQ UniProtKB: Actin-related protein 2/3 complex subunit 3 |
-Macromolecule #4: Actin-related protein 2
| Macromolecule | Name: Actin-related protein 2 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.818711 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDSQGRKVVV CDNGTGFVKC GYAGSNFPEH IFPAIVGRPI IRSTTKVGNI EIKDLMVGDE ASELRSMLEV NYPMENGIVR NWDDMKHLW DYTFGPEKLN IDTRNCKILL TEPPMNPTKN REKIVEVMFE TYQFSGVYVA IQAVLTLYAQ GLLTGVVVDS G DGVTHICP ...String: MDSQGRKVVV CDNGTGFVKC GYAGSNFPEH IFPAIVGRPI IRSTTKVGNI EIKDLMVGDE ASELRSMLEV NYPMENGIVR NWDDMKHLW DYTFGPEKLN IDTRNCKILL TEPPMNPTKN REKIVEVMFE TYQFSGVYVA IQAVLTLYAQ GLLTGVVVDS G DGVTHICP VYEGFSLPHL TRRLDIAGRD ITRYLIKLLL LRGYAFNHSA DFETVRMIKE KLCYVGYNIE QEQKLALETT VL VESYTLP DGRIIKVGGE RFEAPEALFQ PHLINVEGVG VAELLFNTIQ AADIDTRSEF YKHIVLSGGS TMYPGLPSRL ERE LKQLYL ERVLKGDVEK LSKFKIRIED PPRRKHMVFL GGAVLADIMK DKDNFWMTRQ EYQEKGVRVL EKLGVTVR UniProtKB: Actin-related protein 2 |
-Macromolecule #5: Actin-related protein 2/3 complex subunit 4
| Macromolecule | Name: Actin-related protein 2/3 complex subunit 4 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 19.697047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTATLRPYLS AVRATLQAAL CLENFSSQVV ERHNKPEVEV RSSKELLLQP VTISRNEKEK VLIEGSINSV RVSIAVKQAD EIEKILCHK FMRFMMMRAE NFFILRRKPV EGYDISFLIT NFHTEQMYKH KLVDFVIHFM EEIDKEISEM KLSVNARARI V AEEFLKNF UniProtKB: Actin-related protein 2/3 complex subunit 4 |
-Macromolecule #6: Actin-related protein 2/3 complex subunit 5
| Macromolecule | Name: Actin-related protein 2/3 complex subunit 5 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 16.341407 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKNTVSSAR FRKVDVDEYD ENKFVDEEDG GDGQAGPDEG EVDSCLRQGN MTAALQAALK NPPINTKSQA VKDRAGSIVL KVLISFKAN DIEKAVQSLD KNGVDLLMKY IYKGFESPSD NSSAMLLQWH EKALAAGGVG SIVRVLTARK TV UniProtKB: Actin-related protein 2/3 complex subunit 5 |
-Macromolecule #7: Actin-related protein 2/3 complex subunit 1A
| Macromolecule | Name: Actin-related protein 2/3 complex subunit 1A / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.624262 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSLHQFLLEP ITCHAWNRDR TQIALSPNNH EVHIYKKNGS QWVKAHELKE HNGHITGIDW APKSDRIVTC GADRNAYVWS QKDGVWKPT LVILRINRAA TFVKWSPLEN KFAVGSGARL ISVCYFESEN DWWVSKHIKK PIRSTVLSLD WHPNNVLLAA G SCDFKCRV ...String: MSLHQFLLEP ITCHAWNRDR TQIALSPNNH EVHIYKKNGS QWVKAHELKE HNGHITGIDW APKSDRIVTC GADRNAYVWS QKDGVWKPT LVILRINRAA TFVKWSPLEN KFAVGSGARL ISVCYFESEN DWWVSKHIKK PIRSTVLSLD WHPNNVLLAA G SCDFKCRV FSAYIKEVDE KPASTPWGSK MPFGQLMSEF GGSGTGGWVH GVSFSASGSR LAWVSHDSTV SVADASKSVQ VS TLKTEFL PLLSVSFVSE NSVVAAGHDC CPMLFNYDDR GCLTFVSKLD IPKQSIQRNM SAMERFRNMD KRATTEDRNT ALE TLHQNS ITQVSIYEVD KQDCRKFCTT GIDGAMTIWD FKTLESSIQG LRIM UniProtKB: Actin-related protein 2/3 complex subunit 1A |
-Macromolecule #8: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 8 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 59.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 4.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 130973 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)