-Search query
-Search result
Showing 1 - 50 of 85 items for (author: li & zq)

EMDB-18950: 
70S Escherichia coli ribosome with Paenilamicin B2 bound with A- and P-site tRNA.

EMDB-19004: 
70S Escherichia coli ribosome with Paenilamicin B2 bound with hybrid A/P- and hybrid P/E-tRNA.

PDB-8r6c: 
70S Escherichia coli ribosome with Paenilamicin B2 bound with A- and P-site tRNA.

PDB-8r8m: 
70S Escherichia coli ribosome with Paenilamicin B2 bound with hybrid A/P- and hybrid P/E-tRNA.

EMDB-50296: 
70S Escherichia coli ribosome with P-site initiatior tRNA.

PDB-9fbv: 
70S Escherichia coli ribosome with P-site initiatior tRNA.

EMDB-35595: 
Cryo-EM structure of Cas12j-SF05-crRNA-dsDNA complex

EMDB-33797: 
Cryo-EM structure of the EfPiwi (N959K)-piRNA-target ternary complex

EMDB-33800: 
Cryo-EM structure of Hili in complex with piRNA

EMDB-33801: 
Cryo-EM structure of the Mili-piRNA- target ternary complex

EMDB-33809: 
Cryo-EM structure of the EfPiwi(N959K) in complex with piRNA

EMDB-33817: 
Cryo-EM structure of the Mili in complex with piRNA

EMDB-16453: 
SARS-CoV-2 Omicron Variant Spike Trimer in complex with three 17T2 Fabs

EMDB-16473: 
SARS-CoV-2 spike in complex with the 17T2 neutralizing antibody Fab fragment (local refinement of RBD and Fab)

PDB-8c89: 
SARS-CoV-2 spike in complex with the 17T2 neutralizing antibody Fab fragment (local refinement of RBD and Fab)

EMDB-35240: 
Cryo-EM structure of TIR-APAZ/Ago-gRNA-DNA complex
EMDB-40557: 
Cryo-EM structure of designed Influenza HA binder, HA_20, bound to Influenza HA (Strain: Iowa43)

PDB-8sk7: 
Cryo-EM structure of designed Influenza HA binder, HA_20, bound to Influenza HA (Strain: Iowa43)
EMDB-35767: 
Structure of the Mex67-Mtr2-3 heterodimer

EMDB-34638: 
Structure of the Mex67-Mtr2-1 heterodimer
EMDB-34640: 
Structure of Mex67-Mtr2-2 heterodimer
EMDB-34641: 
Structure of Crm1-RanGTP complex

EMDB-34725: 
NPC-trapped pre-60S particle
EMDB-35812: 
Bud20 interacts with CFNC

EMDB-33923: 
Cryo-EM structure of SARS-CoV-2 Omicron spike glycoprotein in complex with three neutralizing nanobody 3-2A2-4

EMDB-25362: 
Cryo-electron tomography structure of membrane-bound EHD4 complex

PDB-7sox: 
Cryo-electron tomography structure of membrane-bound EHD4 complex

EMDB-32438: 
The structure of PldA-PA3488 complex

EMDB-32091: 
Cryo-EM structure of Ams1 bound to the FW domain of Nbr1

EMDB-32653: 
Cryo-EM structure of the inner ring protomer of the Saccharomyces cerevisiae nuclear pore complex

EMDB-32658: 
Cryo-EM structure of the inner ring monomer of the Saccharomyces cerevisiae nuclear pore complex

EMDB-32662: 
Cryo-EM map of the inner ring dimer of the Saccharomyces cerevisiae nuclear pore complex
EMDB-32663: 
Cryo-EM map of the intact inner ring of the Saccharomyces cerevisiae nuclear pore complex

EMDB-32664: 
Cryo-EM map of the whole Saccharomyces cerevisiae nuclear pore complex

EMDB-32643: 
Cryo-EM structure of full-length Nup188

EMDB-32644: 
Cryo-EM structure of full-length Nup188 (multiBody refined N-terminal region)

EMDB-32645: 
Cryo-EM structure of full-length Nup188 (multibody refined C-terminal region)

EMDB-26162: 
Nipah Virus attachment (G) glycoprotein ectodomain in complex with nAH1.3 neutralizing antibody Fab fragment (local refinement of the distal region)

EMDB-26163: 
Nipah Virus attachment (G) glycoprotein ectodomain in complex with nAH1.3 neutralizing antibody Fab fragment (local refinement of the stalk region)

EMDB-26164: 
Nipah Virus attachment (G) glycoprotein ectodomain in complex with nAH1.3 neutralizing antibody Fab fragment (global refinement)

EMDB-31502: 
S protein of SARS-CoV-2 in complex bound with P36-5D2(state2)

EMDB-31503: 
S protein of SARS-CoV-2 in complex bound with P36-5D2 (state1)

EMDB-24607: 
SARS-CoV-2 spike in complex with the S2X58 neutralizing antibody Fab fragment (two receptor-binding domains open)

EMDB-24608: 
SARS-CoV-2 spike in complex with the S2X58 neutralizing antibody Fab fragment (three receptor-binding domains open)
EMDB-20344: 
Polyclonal serum of rabbit C3 post 5th immunization in complex with heterologous Env trimer 45_01dG5 NFL TD 2CC+

EMDB-20259: 
HIV Env BG505 NFL TD+ in complex with antibody E70 fragment antigen binding

EMDB-20260: 
HIV Env 16055 NFL TD 2CC+ in complex with antibody 1C2 fragment antigen binding
EMDB-20273: 
Negative-stain reconstruction of HIV-1 Env BG505 NFL CC+ trimer in complex with rabbit antibody E70 fragment antigen binding
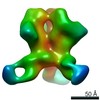
EMDB-20274: 
Negative stain reconstruction of HIV-1 Env 16055 NFL TD 2CC+ trimer in complex with rabbit antibody 1C2 fragment antigen binding
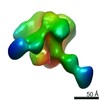
EMDB-20279: 
Polyclonal Fabs from rabbit C3 post 5th immunization in complex with heterologous Env trimer 45_01dH5
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
