+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of TIR-APAZ/Ago-gRNA-DNA complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | a protein complex / ANTIVIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Maribacter polysiphoniae (bacteria) Maribacter polysiphoniae (bacteria) | |||||||||
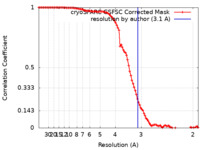
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Zhang H / Deng ZQ / Yu GM / Li XZ / Wang XS | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2023 Journal: Cell Res / Year: 2023Title: Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity. Authors: Xiaoshen Wang / Xuzichao Li / Guimei Yu / Lingling Zhang / Chendi Zhang / Yong Wang / Fumeng Liao / Yanan Wen / Hang Yin / Xiang Liu / Yong Wei / Zhuang Li / Zengqin Deng / Heng Zhang /  Abstract: Nicotinamide adenine dinucleotide (NAD) is a central metabolite in cellular processes. Depletion of NAD has been demonstrated to be a prevalent theme in both prokaryotic and eukaryotic immune ...Nicotinamide adenine dinucleotide (NAD) is a central metabolite in cellular processes. Depletion of NAD has been demonstrated to be a prevalent theme in both prokaryotic and eukaryotic immune responses. Short prokaryotic Argonaute proteins (Agos) are associated with NADase domain-containing proteins (TIR-APAZ or SIR2-APAZ) encoded in the same operon. They confer immunity against mobile genetic elements, such as bacteriophages and plasmids, by inducing NAD depletion upon recognition of target nucleic acids. However, the molecular mechanisms underlying the activation of such prokaryotic NADase/Ago immune systems remain unknown. Here, we report multiple cryo-EM structures of NADase/Ago complexes from two distinct systems (TIR-APAZ/Ago and SIR2-APAZ/Ago). Target DNA binding triggers tetramerization of the TIR-APAZ/Ago complex by a cooperative self-assembly mechanism, while the heterodimeric SIR2-APAZ/Ago complex does not assemble into higher-order oligomers upon target DNA binding. However, the NADase activities of these two systems are unleashed via a similar closed-to-open transition of the catalytic pocket, albeit by different mechanisms. Furthermore, a functionally conserved sensor loop is employed to inspect the guide RNA-target DNA base pairing and facilitate the conformational rearrangement of Ago proteins required for the activation of these two systems. Overall, our study reveals the mechanistic diversity and similarity of Ago protein-associated NADase systems in prokaryotic immune response. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35240.map.gz emd_35240.map.gz | 97.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35240-v30.xml emd-35240-v30.xml emd-35240.xml emd-35240.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35240_fsc.xml emd_35240_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_35240.png emd_35240.png | 140.5 KB | ||
| Others |  emd_35240_half_map_1.map.gz emd_35240_half_map_1.map.gz emd_35240_half_map_2.map.gz emd_35240_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35240 http://ftp.pdbj.org/pub/emdb/structures/EMD-35240 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35240 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35240 | HTTPS FTP |
-Validation report
| Summary document |  emd_35240_validation.pdf.gz emd_35240_validation.pdf.gz | 985.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35240_full_validation.pdf.gz emd_35240_full_validation.pdf.gz | 985.4 KB | Display | |
| Data in XML |  emd_35240_validation.xml.gz emd_35240_validation.xml.gz | 17.7 KB | Display | |
| Data in CIF |  emd_35240_validation.cif.gz emd_35240_validation.cif.gz | 22.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35240 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35240 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35240 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35240 | HTTPS FTP |
-Related structure data
| Related structure data |  8i87MC  8i88C  8in8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35240.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35240.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35240_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35240_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TIR-APAZ/Ago-gRNA-DNA complex
| Entire | Name: TIR-APAZ/Ago-gRNA-DNA complex |
|---|---|
| Components |
|
-Supramolecule #1: TIR-APAZ/Ago-gRNA-DNA complex
| Supramolecule | Name: TIR-APAZ/Ago-gRNA-DNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1, #4, #3, #2 |
|---|---|
| Source (natural) | Organism:  Maribacter polysiphoniae (bacteria) Maribacter polysiphoniae (bacteria) |
-Macromolecule #1: TIR domain-containing protein
| Macromolecule | Name: TIR domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Maribacter polysiphoniae (bacteria) Maribacter polysiphoniae (bacteria) |
| Molecular weight | Theoretical: 53.270594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRNKIFISHA TPDDNDFTRW LALKLIGLGY EVWCDILFLD KGVDFWSNIE KVIREDTCKF LLVSSSYSNQ REGVLKELAV AAKVKKQLK DDKFIIPLAI DEQLSYDDIN IDIVRLNAID FKMSWARGLK DILEAFEKQK VPKEVADASK SNLLYQQIFL H DKSVIEKE ...String: MRNKIFISHA TPDDNDFTRW LALKLIGLGY EVWCDILFLD KGVDFWSNIE KVIREDTCKF LLVSSSYSNQ REGVLKELAV AAKVKKQLK DDKFIIPLAI DEQLSYDDIN IDIVRLNAID FKMSWARGLK DILEAFEKQK VPKEVADASK SNLLYQQIFL H DKSVIEKE EIYDSNWLSI LSFPEELRFH EYNWMLPKRF DVRELTFPAV RYKNYLCTFA WAYDFTYHLP KTETYHKSKT IR IPTEEIL SGSYDSNFIR NAECKRLIVQ LLNKAFELRM KDKEVQEYEM SNKTAYWLEK GKLEKDKFEK TMLVGKQKDK NWH FAISGA SKLYPFPVLM ISSHIFFTAD GKKLIDSSSV QHSSRRRQGK NWWNNTWRTK LLAFIKYLSD DDTSFYLEMG SEEK VFVSN EPVKFKGNVS YNIPEKNTLE EEAELSGFNQ GEDIEELEEL IENLEAE UniProtKB: TIR domain-containing protein |
-Macromolecule #2: Piwi domain-containing protein
| Macromolecule | Name: Piwi domain-containing protein / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Maribacter polysiphoniae (bacteria) Maribacter polysiphoniae (bacteria) |
| Molecular weight | Theoretical: 58.09141 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKELIYIEEP KILFAHGQKC TDARDGLALF GPLNNLYGIK SGVIGTKQGL KIFRDYLDHI QKPIYNSNSI TRPMFPGFEA VFDCKWEST GITFKEVTNE DIGKFLYNSS THKRTYDLVS LFIDKIISAN KNEDENVDVW FVIVPDEIYK YCRPNSVLPK E MVQTKALM ...String: MKELIYIEEP KILFAHGQKC TDARDGLALF GPLNNLYGIK SGVIGTKQGL KIFRDYLDHI QKPIYNSNSI TRPMFPGFEA VFDCKWEST GITFKEVTNE DIGKFLYNSS THKRTYDLVS LFIDKIISAN KNEDENVDVW FVIVPDEIYK YCRPNSVLPK E MVQTKALM SKSKAKSFRY EPSLFPDINI ELKEQEKEAE TYNYDAQFHD QFKARLLKHT IPTQIFREST LAWRDFKNAF GL PIRDFSK IEGHLAWTIS TAAFYKAGGK PWKLSDVRNG VCYLGLVYKK VEKSKNPRNA CCAAQMFLDN GDGTVFKGEV GPW YNPKNG QYHLEPKEAK ALLSQSLQSY KEQIGEYPKE VFIHAKTRFN HQEWDAFLEV TPKETNLVGV TISKTKPLKL YKTE GDYTI LRGNAYVVNE RSAFLWTVGY VPKIQTALSM EVPNPLFIEI NKGEADIKQV LKDILSLTKL NYNACIFADG EPVTL RFAD KIGEILTAST DIKTPPLAFK YYI UniProtKB: Piwi domain-containing protein |
-Macromolecule #3: DNA (5'-D(P*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP...
| Macromolecule | Name: DNA (5'-D(P*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*A)-3') type: dna / ID: 3 / Number of copies: 4 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Maribacter polysiphoniae (bacteria) Maribacter polysiphoniae (bacteria) |
| Molecular weight | Theoretical: 5.692727 KDa |
| Sequence | String: (DT)(DA)(DT)(DA)(DC)(DA)(DA)(DC)(DC)(DT) (DA)(DC)(DT)(DA)(DC)(DC)(DT)(DC)(DA) |
-Macromolecule #4: RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP...
| Macromolecule | Name: RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*A)-3') type: rna / ID: 4 / Number of copies: 4 |
|---|---|
| Source (natural) | Organism:  Maribacter polysiphoniae (bacteria) Maribacter polysiphoniae (bacteria) |
| Molecular weight | Theoretical: 6.160674 KDa |
| Sequence | String: UGAGGUAGUA GGUUGUAUA |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)