-Search query
-Search result
Showing 1 - 50 of 157 items for (author: leo & ys)

EMDB-18180: 
cryoEM structure of SARS-CoV2 Spike trimer in complex with Fab23

PDB-8q5y: 
cryoEM structure of SARS-CoV2 Spike trimer in complex with Fab23

EMDB-19136: 
Thinner is not always better: Optimising cryo lamellae for subtomogram averaging

EMDB-19160: 
Thinner is not always better: Optimising cryo lamellae for subtomogram averaging - Lamella thickness analysis

EMDB-19161: 
Thinner is not always better: Optimising cryo lamellae for subtomogram averaging - Ion-damage layer analysis

EMDB-17171: 
CTE typeI tau filament from Guam ALS/PDC

EMDB-17173: 
CTE typeIII tau filament from Guam ALS/PDC

EMDB-17174: 
CTE typeII tau filament from Guam ALS/PDC

EMDB-17175: 
TMEM106B Fold1-s filament from Guam ALS/PDC

EMDB-17176: 
TMEM106B Fold I-d filament from Guam ALS/PDC

EMDB-17177: 
Ab typeII filament from Guam ALS/PDC

EMDB-17178: 
CTE typeI tau filament from Kii ALS/PDC

EMDB-17179: 
TypeII tau filament from Kii ALS/PDC

EMDB-17180: 
CTE typeIII tau filament

EMDB-17181: 
PHF tau filament from Kii ALS/PDC
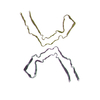
PDB-8ot6: 
CTE typeI tau filament from Guam ALS/PDC

PDB-8ot9: 
CTE typeIII tau filament from Guam ALS/PDC
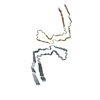
PDB-8otc: 
CTE typeII tau filament from Guam ALS/PDC
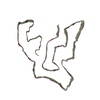
PDB-8otd: 
TMEM106B Fold1-s filament from Guam ALS/PDC

PDB-8ote: 
TMEM106B Fold I-d filament from Guam ALS/PDC

PDB-8otf: 
Ab typeII filament from Guam ALS/PDC
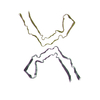
PDB-8otg: 
CTE typeI tau filament from Kii ALS/PDC
PDB-8oth: 
TypeII tau filament from Kii ALS/PDC
PDB-8oti: 
CTE typeIII tau filament
PDB-8otj: 
PHF tau filament from Kii ALS/PDC

EMDB-17819: 
XBB 1.0 RBD bound to P4J15 (Local)
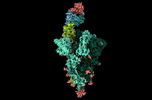
EMDB-17849: 
XBB 1.0 RBD bound to P4J15 (Global)

EMDB-17850: 
SARS-CoV-2 XBB 1.0 closed conformation.

PDB-8pq2: 
XBB 1.0 RBD bound to P4J15 (Local)

PDB-8psd: 
SARS-CoV-2 XBB 1.0 closed conformation.

EMDB-15270: 
SARS Cov2 Spike RBD in complex with Fab47

PDB-8a95: 
SARS Cov2 Spike RBD in complex with Fab47
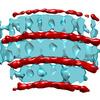
EMDB-16438: 
Toroidal Dps-DNA assembly

EMDB-16439: 
Dps-DNA filament-like assembly

EMDB-15273: 
SARS Cov2 Spike in 1-up conformation complex with Fab47

PDB-8a99: 
SARS Cov2 Spike in 1-up conformation complex with Fab47

EMDB-15269: 
SARS CoV2 Spike in the 2-up state in complex with Fab47

EMDB-15271: 
SARS Cov2 Spike RBD in complex with Fab47

PDB-8a94: 
SARS CoV2 Spike in the 2-up state in complex with Fab47.

PDB-8a96: 
SARS Cov2 Spike RBD in complex with Fab47

EMDB-14815: 
Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Open-ready state, Entire consensus map

EMDB-14816: 
Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Open-ready state, PA focused map

EMDB-14817: 
Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Open-ready state, MD focused map

EMDB-14818: 
Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Resting state, PA focused map

EMDB-14819: 
Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Resting state, MD focused map

EMDB-14820: 
Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Resting state, Entire consensus map

EMDB-14821: 
Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Closed state, Entire consensus map

EMDB-14822: 
Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Closed state, PA focused map

EMDB-14823: 
Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Closed state, MD focused map

EMDB-14824: 
Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Open state, Entire consensus map
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
