[English] 日本語
 Yorodumi
Yorodumi- EMDB-19161: Thinner is not always better: Optimising cryo lamellae for subtom... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Thinner is not always better: Optimising cryo lamellae for subtomogram averaging - Ion-damage layer analysis | |||||||||
 Map data Map data | In-situ ribosome from Dictyostelium discoideum obtained with subtomogram averaging. Ion-damage analysis: 50 nm from lamella surface | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | In-situ Dictyostelium discoideum ribosome / RIBOSOME | |||||||||
| Biological species |  | |||||||||
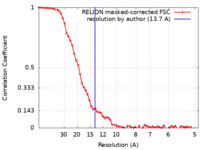
| Method | subtomogram averaging / cryo EM / Resolution: 13.7 Å | |||||||||
 Authors Authors | Tuijtel MW / Cruz-Leon S / Kreysing JP / Welsch S / Hummer G / Beck M / Turonova B | |||||||||
| Funding support |  United States, United States,  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Thinner is not always better: Optimizing cryo-lamellae for subtomogram averaging. Authors: Maarten W Tuijtel / Sergio Cruz-León / Jan Philipp Kreysing / Sonja Welsch / Gerhard Hummer / Martin Beck / Beata Turoňová /  Abstract: Cryo-electron tomography (cryo-ET) is a powerful method to elucidate subcellular architecture and to structurally analyze biomolecules in situ by subtomogram averaging, yet data quality critically ...Cryo-electron tomography (cryo-ET) is a powerful method to elucidate subcellular architecture and to structurally analyze biomolecules in situ by subtomogram averaging, yet data quality critically depends on specimen thickness. Cells that are too thick for transmission imaging can be thinned into lamellae by cryo-focused ion beam (cryo-FIB) milling. Despite being a crucial parameter directly affecting attainable resolution, optimal lamella thickness has not been systematically investigated nor the extent of structural damage caused by gallium ions used for FIB milling. We thus systematically determined how resolution is affected by these parameters. We find that ion-induced damage does not affect regions more than 30 nanometers from either lamella surface and that up to ~180-nanometer lamella thickness does not negatively affect resolution. This shows that there is no need to generate very thin lamellae and lamella thickness can be chosen such that it captures cellular features of interest, thereby opening cryo-ET also for studies of large complexes. #1:  Journal: BiorXiv Journal: BiorXivTitle: Thinner is not always better: Optimising cryo lamellae for subtomogram averaging Authors: Tuijtel MW / Cruz-Leon S / Kreysing JP / Welsch S / Hummer G / Beck M / Turonova B | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19161.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19161.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | In-situ ribosome from Dictyostelium discoideum obtained with subtomogram averaging. Ion-damage analysis: 50 nm from lamella surface | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.446 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Additional map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Half map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
+Half map: In-situ ribosome from Dictyostelium discoideum obtained with subtomogram...
- Sample components
Sample components
-Entire : Ribosome
| Entire | Name: Ribosome |
|---|---|
| Components |
|
-Supramolecule #1: Ribosome
| Supramolecule | Name: Ribosome / type: complex / ID: 1 / Parent: 0 / Details: Ribosome |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 4 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 20 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number real images: 2 / Average exposure time: 0.24 sec. / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 2.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller About Yorodumi
About Yorodumi






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)