+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Dps-DNA filament-like assembly | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Co-crystal / DNA / Dps / biocrystallization / DNA BINDING PROTEIN | |||||||||
| Biological species |  | |||||||||
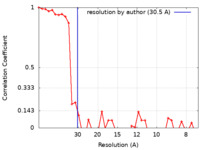
| Method | subtomogram averaging / cryo EM / Resolution: 30.5 Å | |||||||||
 Authors Authors | Chesnokov YM | |||||||||
| Funding support |  Russian Federation, 1 items Russian Federation, 1 items
| |||||||||
 Citation Citation |  Journal: Int J Mol Sci / Year: 2023 Journal: Int J Mol Sci / Year: 2023Title: Morphological Diversity of Dps Complex with Genomic DNA. Authors: Yuri Chesnokov / Roman Kamyshinsky / Andrey Mozhaev / Eleonora Shtykova / Alexander Vasiliev / Ivan Orlov / Liubov Dadinova /  Abstract: In response to adverse environmental factors, cells actively produce Dps proteins which form ordered complexes (biocrystals) with bacterial DNA to protect the genome. The effect of ...In response to adverse environmental factors, cells actively produce Dps proteins which form ordered complexes (biocrystals) with bacterial DNA to protect the genome. The effect of biocrystallization has been described extensively in the scientific literature; furthermore, to date, the structure of the Dps-DNA complex has been established in detail in vitro using plasmid DNA. In the present work, for the first time, Dps complexes with genomic DNA were studied in vitro using cryo-electron tomography. We demonstrate that genomic DNA forms one-dimensional crystals or filament-like assemblies which transform into weakly ordered complexes with triclinic unit cells, similar to what is observed for plasmid DNA. Changing such environmental factors as pH and KCl and MgCl concentrations leads to the formation of cylindrical structures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16439.map.gz emd_16439.map.gz | 878 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16439-v30.xml emd-16439-v30.xml emd-16439.xml emd-16439.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16439_fsc.xml emd_16439_fsc.xml | 4.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_16439.png emd_16439.png | 96.2 KB | ||
| Masks |  emd_16439_msk_1.map emd_16439_msk_1.map | 3.4 MB |  Mask map Mask map | |
| Others |  emd_16439_half_map_1.map.gz emd_16439_half_map_1.map.gz emd_16439_half_map_2.map.gz emd_16439_half_map_2.map.gz | 1.7 MB 1.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16439 http://ftp.pdbj.org/pub/emdb/structures/EMD-16439 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16439 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16439 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16439.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16439.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.7 Å | ||||||||||||||||||||||||||||||||||||
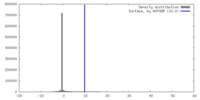
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16439_msk_1.map emd_16439_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
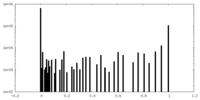
| Density Histograms |
-Half map: #1
| File | emd_16439_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
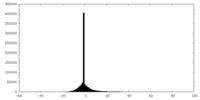
| Density Histograms |
-Half map: #2
| File | emd_16439_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
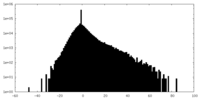
| Density Histograms |
- Sample components
Sample components
-Entire : Dps-DNA assembly
| Entire | Name: Dps-DNA assembly |
|---|---|
| Components |
|
-Supramolecule #1: Dps-DNA assembly
| Supramolecule | Name: Dps-DNA assembly / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 220 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 3.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.026000000000000002 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV / Details: 2.5 s blot. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number real images: 2 / Average exposure time: 2.5 sec. / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 8.0 µm / Calibrated defocus min: 6.0 µm / Calibrated magnification: 37837 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 8.0 µm / Nominal defocus min: 6.0 µm / Nominal magnification: 18000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: HELIUM |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)