+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | TMEM106B Fold I-d filament from Guam ALS/PDC | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TMEM106B / Guam ALS/PDC / PROTEIN FIBRIL | |||||||||
| Function / homology |  Function and homology information Function and homology informationlysosomal protein catabolic process / lysosomal lumen acidification / regulation of lysosome organization / lysosome localization / positive regulation of dendrite development / lysosomal transport / lysosome organization / dendrite morphogenesis / neuron cellular homeostasis / late endosome membrane ...lysosomal protein catabolic process / lysosomal lumen acidification / regulation of lysosome organization / lysosome localization / positive regulation of dendrite development / lysosomal transport / lysosome organization / dendrite morphogenesis / neuron cellular homeostasis / late endosome membrane / ATPase binding / lysosome / endosome / lysosomal membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Qi C / Yang S / Scheres SHW / Goedert M | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Tau filaments from amyotrophic lateral sclerosis/parkinsonism-dementia complex adopt the CTE fold. Authors: Chao Qi / Bert M Verheijen / Yasumasa Kokubo / Yang Shi / Stephan Tetter / Alexey G Murzin / Asa Nakahara / Satoru Morimoto / Marc Vermulst / Ryogen Sasaki / Eleonora Aronica / Yoshifumi ...Authors: Chao Qi / Bert M Verheijen / Yasumasa Kokubo / Yang Shi / Stephan Tetter / Alexey G Murzin / Asa Nakahara / Satoru Morimoto / Marc Vermulst / Ryogen Sasaki / Eleonora Aronica / Yoshifumi Hirokawa / Kiyomitsu Oyanagi / Akiyoshi Kakita / Benjamin Ryskeldi-Falcon / Mari Yoshida / Masato Hasegawa / Sjors H W Scheres / Michel Goedert /     Abstract: The amyotrophic lateral sclerosis/parkinsonism-dementia complex (ALS/PDC) of the island of Guam and the Kii peninsula of Japan is a fatal neurodegenerative disease of unknown cause that is ...The amyotrophic lateral sclerosis/parkinsonism-dementia complex (ALS/PDC) of the island of Guam and the Kii peninsula of Japan is a fatal neurodegenerative disease of unknown cause that is characterized by the presence of abundant filamentous tau inclusions in brains and spinal cords. Here, we used electron cryo-microscopy to determine the structures of tau filaments from the cerebral cortex of three cases of ALS/PDC from Guam and eight cases from Kii, as well as from the spinal cord of two of the Guam cases. Tau filaments had the chronic traumatic encephalopathy (CTE) fold, with variable amounts of Type I and Type II filaments. Paired helical tau filaments were also found in three Kii cases and tau filaments with the corticobasal degeneration fold in one Kii case. We identified a new Type III CTE tau filament, where protofilaments pack against each other in an antiparallel fashion. ALS/PDC is the third known tauopathy with CTE-type filaments and abundant tau inclusions in cortical layers II/III, the others being CTE and subacute sclerosing panencephalitis. Because these tauopathies are believed to have environmental causes, our findings support the hypothesis that ALS/PDC is caused by exogenous factors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17176.map.gz emd_17176.map.gz | 58.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17176-v30.xml emd-17176-v30.xml emd-17176.xml emd-17176.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17176_fsc.xml emd_17176_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_17176.png emd_17176.png | 51.1 KB | ||
| Masks |  emd_17176_msk_1.map emd_17176_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17176.cif.gz emd-17176.cif.gz | 5.2 KB | ||
| Others |  emd_17176_half_map_1.map.gz emd_17176_half_map_1.map.gz emd_17176_half_map_2.map.gz emd_17176_half_map_2.map.gz | 192.1 MB 192.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17176 http://ftp.pdbj.org/pub/emdb/structures/EMD-17176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17176 | HTTPS FTP |
-Related structure data
| Related structure data |  8oteMC  8ot6C  8ot9C  8otcC  8otdC  8otfC  8otgC  8othC  8otiC  8otjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17176.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17176.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||||||||||||||||||
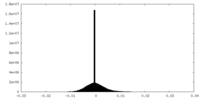
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17176_msk_1.map emd_17176_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17176_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17176_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TMEM106B Fold1-d filaments from Guam ALS/PDC
| Entire | Name: TMEM106B Fold1-d filaments from Guam ALS/PDC |
|---|---|
| Components |
|
-Supramolecule #1: TMEM106B Fold1-d filaments from Guam ALS/PDC
| Supramolecule | Name: TMEM106B Fold1-d filaments from Guam ALS/PDC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Transmembrane protein 106B
| Macromolecule | Name: Transmembrane protein 106B / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.142295 KDa |
| Sequence | String: MGKSLSHLPL HSSKEDAYDG VTSENMRNGL VNSEVHNEDG RNGDVSQFPY VEFTGRDSVT CPTCQGTGRI PRGQENQLVA LIPYSDQRL RPRRTKLYVM ASVFVCLLLS GLAVFFLFPR SIDVKYIGVK SAYVSYDVQK RTIYLNITNT LNITNNNYYS V EVENITAQ ...String: MGKSLSHLPL HSSKEDAYDG VTSENMRNGL VNSEVHNEDG RNGDVSQFPY VEFTGRDSVT CPTCQGTGRI PRGQENQLVA LIPYSDQRL RPRRTKLYVM ASVFVCLLLS GLAVFFLFPR SIDVKYIGVK SAYVSYDVQK RTIYLNITNT LNITNNNYYS V EVENITAQ VQFSKTVIGK ARLNNISIIG PLDMKQIDYT VPTVIAEEMS YMYDFCTLIS IKVHNIVLMM QVTVTTTYFG HS EQISQER YQYVDCGRNT TYQLGQSEYL NVLQPQQ UniProtKB: Transmembrane protein 106B |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)