-Search query
-Search result
Showing all 39 items for (author: lei & yf)

EMDB-64077: 
Cryo-EM structure of SARS-CoV-2 KP.2 spike RBD in complex with ACE2
Method: single particle / : Jin XH, Sun L

EMDB-64078: 
Cryo-EM structure of SARS-CoV-2 KP.2 spike in complex with ACE2
Method: single particle / : Jin XH, Sun L

EMDB-39688: 
BA.2.86 RBD protein in complex with ACE2.
Method: single particle / : Wang YJ, Zhang X, Sun L

EMDB-39689: 
Structure of BA.2.86 spike protein in complex with ACE2.
Method: single particle / : Wang YJ, Zang X, Sun L

EMDB-39690: 
Structure of JN.1 RBD protein in complex with ACE2.
Method: single particle / : Wang YJ, Zhang X, Sun L

EMDB-39691: 
The JN.1 spike protein (S) in complex with ACE2.
Method: single particle / : Wang YJ, Zhang X, Sun L
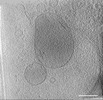
EMDB-39012: 
Representative tomogram of primary glioblastoma stem cell with circular inter-mitochondrial junctions.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39015: 
Representative tomogram of microglia cell with nanotunnel-like structures resembling mitochondrial fission.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39019: 
Representative tomogram of glioblastoma cell with nanotunnel-like structure and inter-mitochondrial junction.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39021: 
Representative tomogram of normal human astrocyte with nanotunnel-like structure which is an extension of the mitochondrial outer membrane.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39023: 
Representative tomogram of primary glioblastoma differentiated cell with parallel inter-mitochondrial junction.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39024: 
Representative tomogram of primary glioblastoma stem cell with clustered mitochondria bearing various long-short axis ratios.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-29943: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Yang D, Sheng C, Jinpeng S

EMDB-29944: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Jiangqian M, Sheng C, Jinpeng S

EMDB-29945: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Jianqiang M, Jinpeng S

EMDB-29946: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 3 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Yang D, Shiyi G, Jinpeng S

EMDB-29935: 
Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein
Method: single particle / : Huang SM, Xiong MY, Liu L, Mu J, Sheng C, Sun J

EMDB-29940: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Huang SM, Xiong MY, Liu L, Mu J, Sheng C, Sun J

EMDB-35705: 
Cryo-EM structure of the DMCHA-bound mTAAR9-Gs complex
Method: single particle / : Sun JP, Li Q, Yang F, Xu YF, Guo LL, Lian S, Zhang MH, Rong NK

EMDB-35761: 
Cryo-EM structure of the PEA-bound mTAAR9-Golf complex
Method: single particle / : Sun JP, Li Q, Yang F, Xu YF, Guo LL, Lian S, Zhang MH, Rong NK

EMDB-35762: 
Cryo-EM structure of the SPE-bound mTAAR9-Gs complex
Method: single particle / : Sun JP, Li Q, Yang F, Xu YF, Guo LL, Lian S, Zhang MH, Rong NK

EMDB-35763: 
Cryo-EM structure of the PEA-bound mTAAR9-Gs complex
Method: single particle / : Sun JP, Li Q, Yang F, Xu YF, Guo LL, Lian S, Zhang MH, Rong NK

EMDB-35764: 
Cryo-EM structure of the CAD-bound mTAAR9-Gs complex
Method: single particle / : Sun JP, Li Q, Yang F, Xu YF, Guo LL, Lian S, Zhang MH, Rong NK

EMDB-35765: 
Cryo-EM structure of the SPE-mTAAR9 complex
Method: single particle / : Sun JP, Li Q, Yang F, Xu YF, Guo LL, Lian S, Zhang MH, Rong NK

EMDB-35771: 
Cryo-EM structure of the PEA-bound mTAAR9 complex
Method: single particle / : Sun JP, Li Q, Yang F, Xu YF, Guo LL, Lian S, Zhang MH, Rong NK

EMDB-31542: 
SARS-COV-2 Spike RBDMACSp6 binding to hACE2
Method: single particle / : Wang X, Cao L

EMDB-31543: 
SARS-COV-2 Spike RBDMACSp25 binding to hACE2
Method: single particle / : Wang X, Cao L

EMDB-31544: 
SARS-COV-2 Spike RBDMACSp36 binding to hACE2
Method: single particle / : Wang X, Cao L

EMDB-31546: 
SARS-COV-2 Spike RBDMACSp36 binding to mACE2
Method: single particle / : Wang X, Cao L

EMDB-30646: 
Structure of Calcium-Sensing Receptor in an inactive state
Method: single particle / : Wen TL, Yang X, Shen YQ

EMDB-22493: 
Cryo-EM structure of SKF-81297-bound dopamine receptor 1 in complex with Gs protein
Method: single particle / : Zhuang Y, Xu P

EMDB-22509: 
Cryo-EM structure of SKF-83959-bound dopamine receptor 1 in complex with Gs protein
Method: single particle / : Zhuang Y, Xu P

EMDB-22510: 
Cryo-EM structure of apomorphine-bound dopamine receptor 1 in complex with Gs protein
Method: single particle / : Zhuang Y, Xu P

EMDB-22511: 
Cryo-EM structure of Bromocriptine-bound dopamine receptor 2 in complex with Gi protein
Method: single particle / : Zhuang Y, Xu P

EMDB-30702: 
S-2H2-F3a structure, two RBDs are up and one RBD is down, each RBD binds with a 2H2 Fab.
Method: single particle / : Cong Y, Wang YF

EMDB-30703: 
S-2H2-F1 structure, one RBD is up and two RBDs are down, only up RBD binds with a 2H2 Fab
Method: single particle / : Cong Y, Wang YF

EMDB-30704: 
S-2H2-F2 structure, two RBDs are up and one RBD is down, each up RBD binds with a 2H2 Fab.
Method: single particle / : Cong Y, Wang YF

EMDB-30705: 
S-2H2-F3b structure, three RBDs are up and each RBD binds with a 2H2 Fab.
Method: single particle / : Cong Y, Wang YF
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
