-Search query
-Search result
Showing 1 - 50 of 3,406 items for (author: lee & l)
EMDB-42603: 
Human p97/VCP structure with a triazole inhibitor (NSC799462/hexamer)
EMDB-42625: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC804515)
EMDB-42626: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC819701/up)
EMDB-42627: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC819701/down)
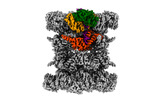
EMDB-44748: 
Human p97/VCP structure with a triazole inhibitor (NSC799462/dodecamer)
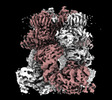
EMDB-44255: 
Cryo-EM structure of the mouse TRPM8 channel in the ligand-free desensitized state

EMDB-44256: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014

EMDB-44257: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist AMG2850
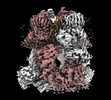
EMDB-44258: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist AMTB
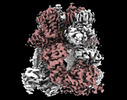
EMDB-44259: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014 and the cooling agonist C3
EMDB-44260: 
Cryo-EM structure of the avian great tit TRPM8 channel in complex with the antagonist TC-I 2014

EMDB-44261: 
Cryo-EM structure of the mouse TRPM8 channel in complex with PI(4,5)P2 and Ca2+
EMDB-44262: 
Cryo-EM structure of the mouse TRPM8 channel in complex with Ca2+ in the absence of PI(4,5)P2

EMDB-37571: 
cryo-EM structure of alligator haemoglobin in carbonmonoxy form

EMDB-37572: 
cryo-EM structure of alligator haemoglobin in oxy form

EMDB-37573: 
cryo-EM structure of alligator haemoglobin in deoxy form

EMDB-37574: 
cryo-EM structure of human haemoglobin in carbonmonoxy form

EMDB-37575: 
cryo-EM structure of human haemoglobin in oxy form

EMDB-37576: 
cryo-EM structure of human haemoglobin in deoxy form

PDB-8wix: 
cryo-EM structure of alligator haemoglobin in carbonmonoxy form

PDB-8wiy: 
cryo-EM structure of alligator haemoglobin in oxy form

PDB-8wiz: 
cryo-EM structure of alligator haemoglobin in deoxy form

PDB-8wj0: 
cryo-EM structure of human haemoglobin in carbonmonoxy form

PDB-8wj1: 
cryo-EM structure of human haemoglobin in oxy form

PDB-8wj2: 
cryo-EM structure of human haemoglobin in deoxy form

EMDB-39212: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH8.0 (3.23A)

EMDB-39213: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH6.5 (2.82A)

EMDB-39214: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH5.0 (3.52A)

EMDB-39215: 
Cryo-EM structure of Dragon Grouper nervous necrosis virion at pH6.5 (3.12A)

EMDB-39217: 
Cryo-EM structure of Dragon Grouper nervous necrosis virion at pH5.0 (4.36A)

PDB-8yf6: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH8.0 (3.23A)

PDB-8yf7: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH6.5 (2.82A)

PDB-8yf8: 
Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH5.0 (3.52A)

PDB-8yf9: 
Cryo-EM structure of Dragon Grouper nervous necrosis virion at pH6.5 (3.12A)

EMDB-18378: 
Focused map for CSA-DDB1-DDA1 (map 2)
EMDB-41485: 
Subtomogram averaged consensus structure of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes
EMDB-41486: 
Subtomogram averaged decoding-1 state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-41487: 
Subtomogram averaged decoding-2 state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-41488: 
Subtomogram averaged classical iPRE state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes
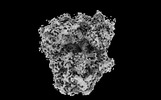
EMDB-41489: 
Subtomogram averaged rotated-1 PRE state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-41490: 
Subtomogram averaged rotated-2 PRE state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-41491: 
Subtomogram averaged translocation state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes
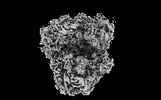
EMDB-41492: 
Subtomogram averaged POST state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-41493: 
Subtomogram averaged unloaded state of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes
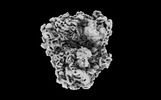
EMDB-41494: 
Subtomogram averaged non-rotated 80S ribosome of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes

EMDB-18380: 
Focused map for UVSSA(VHS)-CSA-DDB1(BPA/BPC)-DDA1

EMDB-18377: 
Focused map for CSA-DDB1-DDA1

EMDB-18398: 
CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1

EMDB-18413: 
Consensus map of UVSSA(VHS)-CSA-DDB1-DDA1

PDB-8qh5: 
CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
