+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of alligator haemoglobin in oxy form | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Haemoglobin / OXYGEN BINDING | |||||||||
| Function / homology |  Function and homology information Function and homology informationhaptoglobin binding / organic acid binding / haptoglobin-hemoglobin complex / hemoglobin complex / oxygen carrier activity / hydrogen peroxide catabolic process / peroxidase activity / oxygen binding / blood microparticle / iron ion binding ...haptoglobin binding / organic acid binding / haptoglobin-hemoglobin complex / hemoglobin complex / oxygen carrier activity / hydrogen peroxide catabolic process / peroxidase activity / oxygen binding / blood microparticle / iron ion binding / heme binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Alligator mississippiensis (American alligator) Alligator mississippiensis (American alligator) | |||||||||
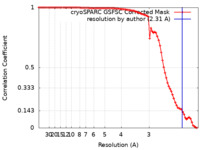
| Method | single particle reconstruction / cryo EM / Resolution: 2.31 Å | |||||||||
 Authors Authors | Takahashi K / Lee Y / Fago A / Bautista NM / Kawamoto A / Kurisu G / Storz J / Nishizawa T / Tame JRH | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM. Authors: Katsuya Takahashi / Yongchan Lee / Angela Fago / Naim M Bautista / Jay F Storz / Akihiro Kawamoto / Genji Kurisu / Tomohiro Nishizawa / Jeremy R H Tame /    Abstract: The principal effect controlling the oxygen affinity of vertebrate haemoglobins (Hbs) is the allosteric switch between R and T forms with relatively high and low oxygen affinity respectively. ...The principal effect controlling the oxygen affinity of vertebrate haemoglobins (Hbs) is the allosteric switch between R and T forms with relatively high and low oxygen affinity respectively. Uniquely among jawed vertebrates, crocodilians possess Hb that shows a profound drop in oxygen affinity in the presence of bicarbonate ions. This allows them to stay underwater for extended periods by consuming almost all the oxygen present in the blood-stream, as metabolism releases carbon dioxide, whose conversion to bicarbonate and hydrogen ions is catalysed by carbonic anhydrase. Despite the apparent universal utility of bicarbonate as an allosteric regulator of Hb, this property evolved only in crocodilians. We report here the molecular structures of both human and a crocodilian Hb in the deoxy and liganded states, solved by cryo-electron microscopy. We reveal the precise interactions between two bicarbonate ions and the crocodilian protein at symmetry-related sites found only in the T state. No other known effector of vertebrate Hbs binds anywhere near these sites. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37572.map.gz emd_37572.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37572-v30.xml emd-37572-v30.xml emd-37572.xml emd-37572.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37572_fsc.xml emd_37572_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_37572.png emd_37572.png | 112.3 KB | ||
| Masks |  emd_37572_msk_1.map emd_37572_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-37572.cif.gz emd-37572.cif.gz | 6.5 KB | ||
| Others |  emd_37572_half_map_1.map.gz emd_37572_half_map_1.map.gz emd_37572_half_map_2.map.gz emd_37572_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37572 http://ftp.pdbj.org/pub/emdb/structures/EMD-37572 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37572 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37572 | HTTPS FTP |
-Related structure data
| Related structure data |  8wiyMC  8wixC  8wizC  8wj0C  8wj1C  8wj2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37572.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37572.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_37572_msk_1.map emd_37572_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_37572_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_37572_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : alligator haemoglobin in carboxy form
| Entire | Name: alligator haemoglobin in carboxy form |
|---|---|
| Components |
|
-Supramolecule #1: alligator haemoglobin in carboxy form
| Supramolecule | Name: alligator haemoglobin in carboxy form / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Alligator mississippiensis (American alligator) Alligator mississippiensis (American alligator) |
| Molecular weight | Theoretical: 100 KDa |
-Macromolecule #1: Hemoglobin subunit alpha
| Macromolecule | Name: Hemoglobin subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Alligator mississippiensis (American alligator) Alligator mississippiensis (American alligator) |
| Molecular weight | Theoretical: 15.891368 KDa |
| Sequence | String: MVLSMEDKSN VKAIWGKASG HLEEYGAEAL ERMFCAYPQT KIYFPHFDMS HNSAQIRAHG KKVFSALHEA VNHIDDLPGA LCRLSELHA HSLRVDPVNF KFLAHCVLVV FAIHHPSALS PEIHASLDKF LCAVSAVLTS KYR UniProtKB: Hemoglobin subunit alpha |
-Macromolecule #2: Hemoglobin subunit beta
| Macromolecule | Name: Hemoglobin subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Alligator mississippiensis (American alligator) Alligator mississippiensis (American alligator) |
| Molecular weight | Theoretical: 16.641213 KDa |
| Sequence | String: ASFDAHERKF IVDLWAKVDV AQCGADALSR MLIVYPWKRR YFEHFGKMCN AHDILHNSKV QEHGKKVLAS FGEAVKHLDN IKGHFANLS KLHCEKFHVD PENFKLLGDI IIIVLAAHHP EDFSVECHAA FQKLVRQVAA ALAAEYH UniProtKB: Hemoglobin subunit beta |
-Macromolecule #3: PROTOPORPHYRIN IX CONTAINING FE
| Macromolecule | Name: PROTOPORPHYRIN IX CONTAINING FE / type: ligand / ID: 3 / Number of copies: 2 / Formula: HEM |
|---|---|
| Molecular weight | Theoretical: 616.487 Da |
| Chemical component information |  ChemComp-HEM: |
-Macromolecule #4: OXYGEN MOLECULE
| Macromolecule | Name: OXYGEN MOLECULE / type: ligand / ID: 4 / Number of copies: 2 / Formula: OXY |
|---|---|
| Molecular weight | Theoretical: 31.999 Da |
| Chemical component information |  ChemComp-O2: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 208 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 15 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Output model |  PDB-8wiy: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)