-Search query
-Search result
Showing 1 - 50 of 65 items for (author: krahn & j)

EMDB-42979: 
Engaged conformation of the human mitochondrial DNA polymerase gamma bound to DNA

EMDB-42980: 
Human mitochondrial DNA polymerase gamma bound to a replication fork in an open conformation

EMDB-42982: 
Human mitochondrial DNA polymerase catalytic subunit, PolG, in an APO conformation

EMDB-42984: 
Active conformation of DNA polymerase gamma bound to DNA

PDB-8v54: 
Engaged conformation of the human mitochondrial DNA polymerase gamma bound to DNA

PDB-8v55: 
Human mitochondrial DNA polymerase gamma bound to a replication fork in an open conformation

PDB-8v5d: 
Human mitochondrial DNA polymerase catalytic subunit, PolG, in an APO conformation

PDB-8v5r: 
Active conformation of DNA polymerase gamma bound to DNA
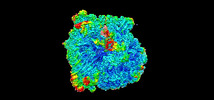
EMDB-42455: 
Candidatus Methanomethylophilus alvus tRNAPyl in A-site of ribosome

PDB-8upt: 
Candidatus Methanomethylophilus alvus tRNAPyl in A-site of ribosome

EMDB-29426: 
Chaetomium thermophilum SETX - NPPC internal deletion

EMDB-29439: 
Chaetomium thermophilum SETX (Full-length)

EMDB-29440: 
SETX-Sen1N domain

PDB-8fth: 
Chaetomium thermophilum SETX - NPPC internal deletion

PDB-8ftk: 
Chaetomium thermophilum SETX (Full-length)

EMDB-28755: 
Human tRNA Splicing Endonuclease Complex bound to 2'F-tRNA-Arg

EMDB-26856: 
Human tRNA Splicing Endonuclease Complex bound to pre-tRNA-ARG

PDB-7uxa: 
Human tRNA Splicing Endonuclease Complex bound to pre-tRNA-ARG

EMDB-26831: 
Human Rix1 sub-complex scaffold

PDB-7uwf: 
Human Rix1 sub-complex scaffold

EMDB-26360: 
Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2

EMDB-26361: 
Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and dGTP

EMDB-26362: 
Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and GTP

PDB-7u65: 
Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2

PDB-7u66: 
Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and dGTP

PDB-7u67: 
Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and GTP
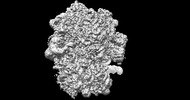
EMDB-26705: 
allo-tRNAUTu1 in the A, P, and E sites of the E. coli ribosome
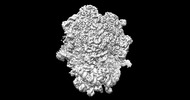
EMDB-26713: 
allo-tRNAUTu1A in the A site of the E. coli ribosome
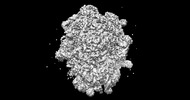
EMDB-26714: 
allo-tRNAUTu1A in the P site of the E. coli ribosome

PDB-7ur5: 
allo-tRNAUTu1 in the A, P, and E sites of the E. coli ribosome

PDB-7uri: 
allo-tRNAUTu1A in the A site of the E. coli ribosome

PDB-7urm: 
allo-tRNAUTu1A in the P site of the E. coli ribosome

EMDB-25743: 
Octameric Human Twinkle Helicase Clinical Variant W315L

EMDB-25744: 
Heptameric Human Twinkle Helicase Clinical Variant W315L

EMDB-25746: 
Octameric Twinkle Helicase Clinical Variant W315L, local refinement

PDB-7t8b: 
Octameric Human Twinkle Helicase Clinical Variant W315L

PDB-7t8c: 
Heptameric Human Twinkle Helicase Clinical Variant W315L

EMDB-25915: 
SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA

EMDB-26073: 
SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA

PDB-7tj2: 
SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA

PDB-7tqv: 
SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA

EMDB-25474: 
CryoEM structure of the N-Terminal deleted Rix7 AAA-ATPase

EMDB-25582: 
CryoEM structure of the crosslinked Rix7 AAA-ATPase

EMDB-25659: 
CryoEM structure of the Rix7 D2 Walker B mutant

PDB-7swl: 
CryoEM structure of the N-terminal-deleted Rix7 AAA-ATPase

PDB-7t0v: 
CryoEM structure of the crosslinked Rix7 AAA-ATPase

PDB-7t3i: 
CryoEM structure of the Rix7 D2 Walker B mutant

EMDB-24137: 
SARS-CoV-2 Nsp15 endoribonuclease pre-cleavage state

PDB-7n33: 
SARS-CoV-2 Nsp15 endoribonuclease pre-cleavage state

EMDB-24101: 
SARS-CoV-2 Nsp15 endoribonuclease post-cleavage state
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
