-Search query
-Search result
Showing 1 - 50 of 147 items for (author: joseph & aj)

EMDB-19822: 
Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/+bromosterol (DOPC, DOPE, DOPS, bromo-ergosterol, PI(4,5)P2 35:20:20:15:10)

EMDB-18307: 
Native eisosome lattice bound to plasma membrane microdomain

EMDB-18308: 
Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture -PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol 30:20:20:30)

EMDB-18309: 
Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/-sterol (DOPC, DOPE, DOPS, PI(4,5)P2 50:20:20:10)

EMDB-18310: 
Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol, PI(4,5)P2 35:20:20:15:10)

EMDB-18311: 
Compact state - Native eisosome lattice bound to plasma membrane microdomain

EMDB-18312: 
Stretched state - Native eisosome lattice bound to plasma membrane microdomain

PDB-8qb7: 
Pil1 in native eisosome lattice bound to plasma membrane microdomain

PDB-8qb8: 
Lsp1 in native eisosome lattice bound to plasma membrane microdomain

PDB-8qb9: 
Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture -PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol 30:20:20:30)

PDB-8qbb: 
Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/-sterol (DOPC, DOPE, DOPS, PI(4,5)P2 50:20:20:10)

PDB-8qbd: 
Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol, PI(4,5)P2 35:20:20:15:10)

PDB-8qbe: 
Compact state - Pil1 in native eisosome lattice bound to plasma membrane microdomain

PDB-8qbf: 
Compact state - Pil1 dimer with lipid headgroups fitted in native eisosome lattice bound to plasma membrane microdomain

PDB-8qbg: 
Stretched state - Pil1 in native eisosome lattice bound to plasma membrane microdomain

EMDB-17295: 
Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation

PDB-8oyt: 
Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation

EMDB-17296: 
Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '2 up 1 down' RBD conformation

PDB-8oyu: 
Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '2 up 1 down' RBD conformation

EMDB-43753: 
Yeast U1 snRNP with humanized U1C Zinc-Finger domain

EMDB-43542: 
ELIC5 with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc in open conformation

EMDB-34530: 
Membrane protein A

EMDB-34531: 
Membrane protein B

EMDB-35713: 
Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc

PDB-8h86: 
Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc

PDB-8h87: 
Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc

PDB-8iu0: 
Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc

EMDB-26855: 
Arabidopsis DDM1 bound to nucleosome (H2A.W, H2B, H3.3, H4, with 147 bp DNA)

PDB-7ux9: 
Arabidopsis DDM1 bound to nucleosome (H2A.W, H2B, H3.3, H4, with 147 bp DNA)

EMDB-27943: 
9H2 Fab-poliovirus 1 complex
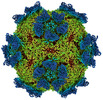
EMDB-27947: 
9H2 Fab-Sabin poliovirus 3 complex

EMDB-27948: 
9H2 Fab-poliovirus 2 complex
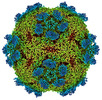
EMDB-27949: 
9H2 Fab-Sabin poliovirus 3 complex
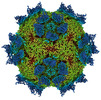
EMDB-27950: 
9H2 Fab-Sabin poliovirus 2 complex
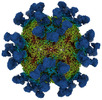
EMDB-27951: 
9H2 Fab-Sabin poliovirus 1 complex
EMDB-40557: 
Cryo-EM structure of designed Influenza HA binder, HA_20, bound to Influenza HA (Strain: Iowa43)
EMDB-16450: 
Production of antigenically stable enterovirus A71 virus-like particles in Pichia pastoris as a vaccine candidate.

EMDB-25612: 
CryoEM structure of mu-opioid receptor - Gi protein complex bound to mitragynine pseudoindoxyl (MP)

EMDB-25613: 
CryoEM structure of mu-opioid receptor - Gi protein complex bound to lofentanil (LFT)

PDB-7t2g: 
CryoEM structure of mu-opioid receptor - Gi protein complex bound to mitragynine pseudoindoxyl (MP)

PDB-7t2h: 
CryoEM structure of mu-opioid receptor - Gi protein complex bound to lofentanil (LFT)

EMDB-27215: 
ELIC apo in POPC nanodisc

EMDB-27216: 
ELIC with cysteamine in POPC nanodisc

EMDB-27217: 
ELIC apo in 2:1:1 POPC:POPE:POPG nanodisc

EMDB-27218: 
ELIC with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc

EMDB-27219: 
ELIC3 with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc

EMDB-26831: 
Human Rix1 sub-complex scaffold

EMDB-25660: 
Cryo-EM structure of Csy-AcrIF24
EMDB-25661: 
Cryo-EM structure of Csy-AcrIF24 dimer

EMDB-25662: 
Cryo-EM structure of Csy-AcrIF24-DNA dimer
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
