[English] 日本語
 Yorodumi
Yorodumi- EMDB-19822: Helical reconstruction of yeast eisosome protein Pil1 bound to me... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/+bromosterol (DOPC, DOPE, DOPS, bromo-ergosterol, PI(4,5)P2 35:20:20:15:10) | ||||||||||||
 Map data Map data | Sharpened helical map 2b PIP2/ bromosterol (D1, rise 5.338, twist 133.559) | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | BAR domain / lipid reconstitution / membrane microdomain / LIPID BINDING PROTEIN | ||||||||||||
| Function / homology | Eisosome component PIL1/LSP1 / Eisosome component PIL1 / AH/BAR domain superfamily / Sphingolipid long chain base-responsive protein PIL1 Function and homology information Function and homology information | ||||||||||||
| Biological species |  | ||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.85 Å | ||||||||||||
 Authors Authors | Kefauver JM / Zou L / Desfosses A / Loewith RJ | ||||||||||||
| Funding support | European Union,  Switzerland, 3 items Switzerland, 3 items
| ||||||||||||
 Citation Citation | Journal: Acta Crystallogr D Struct Biol / Year: 2018 Title: Real-space refinement in PHENIX for cryo-EM and crystallography. Authors: Pavel V Afonine / Billy K Poon / Randy J Read / Oleg V Sobolev / Thomas C Terwilliger / Alexandre Urzhumtsev / Paul D Adams /    Abstract: This article describes the implementation of real-space refinement in the phenix.real_space_refine program from the PHENIX suite. The use of a simplified refinement target function enables very fast ...This article describes the implementation of real-space refinement in the phenix.real_space_refine program from the PHENIX suite. The use of a simplified refinement target function enables very fast calculation, which in turn makes it possible to identify optimal data-restraint weights as part of routine refinements with little runtime cost. Refinement of atomic models against low-resolution data benefits from the inclusion of as much additional information as is available. In addition to standard restraints on covalent geometry, phenix.real_space_refine makes use of extra information such as secondary-structure and rotamer-specific restraints, as well as restraints or constraints on internal molecular symmetry. The re-refinement of 385 cryo-EM-derived models available in the Protein Data Bank at resolutions of 6 Å or better shows significant improvement of the models and of the fit of these models to the target maps. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19822.map.gz emd_19822.map.gz | 1.5 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19822-v30.xml emd-19822-v30.xml emd-19822.xml emd-19822.xml | 25.8 KB 25.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_19822.png emd_19822.png | 69.3 KB | ||
| Masks |  emd_19822_msk_1.map emd_19822_msk_1.map | 1.6 GB |  Mask map Mask map | |
| Filedesc metadata |  emd-19822.cif.gz emd-19822.cif.gz | 5.9 KB | ||
| Others |  emd_19822_additional_1.map.gz emd_19822_additional_1.map.gz emd_19822_additional_2.map.gz emd_19822_additional_2.map.gz emd_19822_additional_3.map.gz emd_19822_additional_3.map.gz emd_19822_additional_4.map.gz emd_19822_additional_4.map.gz emd_19822_half_map_1.map.gz emd_19822_half_map_1.map.gz emd_19822_half_map_2.map.gz emd_19822_half_map_2.map.gz | 813.5 MB 805.7 MB 816.7 MB 809.6 MB 1.5 GB 1.5 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19822 http://ftp.pdbj.org/pub/emdb/structures/EMD-19822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19822 | HTTPS FTP |
-Related structure data
| Related structure data |  8qb7C  8qb8C  8qb9C  8qbbC  8qbdC  8qbeC  8qbfC  8qbgC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19822.map.gz / Format: CCP4 / Size: 1.6 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19822.map.gz / Format: CCP4 / Size: 1.6 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened helical map 2b PIP2/ bromosterol (D1, rise 5.338, twist 133.559) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size |
| ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_19822_msk_1.map emd_19822_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
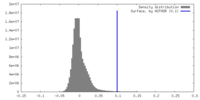
| Density Histograms |
-Additional map: Unsharpened helical map 2b PIP2/ bromosterol (D1, rise...
| File | emd_19822_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened helical map 2b PIP2/ bromosterol (D1, rise 5.338, twist 133.559) | ||||||||||||
| Projections & Slices |
| ||||||||||||
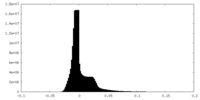
| Density Histograms |
-Additional map: Unsharpened helical map 1b PIP2/ bromosterol (D4, rise...
| File | emd_19822_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened helical map 1b PIP2/ bromosterol (D4, rise 20.64, twist 219.047) | ||||||||||||
| Projections & Slices |
| ||||||||||||
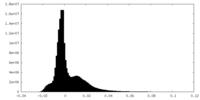
| Density Histograms |
-Additional map: Unsharpened helical map 8b PIP2/ bromosterol (D2, rise...
| File | emd_19822_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened helical map 8b PIP2/ bromosterol (D2, rise 11.025, twist 42.306) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened helical map 9b PIP2/ bromosterol (D1, rise...
| File | emd_19822_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened helical map 9b PIP2/ bromosterol (D1, rise 5.571, twist 152.247) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Helical map 2b PIP2/ bromosterol - half map A
| File | emd_19822_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical map 2b PIP2/ bromosterol - half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Helical assembly of recombinant Pil1 protein tubulating +PIP2/+br...
| Entire | Name: Helical assembly of recombinant Pil1 protein tubulating +PIP2/+bromosterol lipid mixture (DOPC, DOPE, DOPS, bromo-ergosterol, PI(4,5)P2 35:20:20:15:10) |
|---|---|
| Components |
|
-Supramolecule #1: Helical assembly of recombinant Pil1 protein tubulating +PIP2/+br...
| Supramolecule | Name: Helical assembly of recombinant Pil1 protein tubulating +PIP2/+bromosterol lipid mixture (DOPC, DOPE, DOPS, bromo-ergosterol, PI(4,5)P2 35:20:20:15:10) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Sphingolipid long chain base-responsive protein PIL1
| Macromolecule | Name: Sphingolipid long chain base-responsive protein PIL1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MHRTYSLRNS RAPTASQLQN PPPPPSTTKG RFFGKGGLAY SFRRSAAGAF GPELSRKLSQ LVKIEKNVLR SMELTANERR DAAKQLSIW GLENDDDVSD ITDKLGVLIY EVSELDDQFI DRYDQYRLTL KSIRDIEGSV QPSRDRKDKI TDKIAYLKYK D PQSPKIEV ...String: MHRTYSLRNS RAPTASQLQN PPPPPSTTKG RFFGKGGLAY SFRRSAAGAF GPELSRKLSQ LVKIEKNVLR SMELTANERR DAAKQLSIW GLENDDDVSD ITDKLGVLIY EVSELDDQFI DRYDQYRLTL KSIRDIEGSV QPSRDRKDKI TDKIAYLKYK D PQSPKIEV LEQELVRAEA ESLVAEAQLS NITRSKLRAA FNYQFDSIIE HSEKIALIAG YGKALLELLD DSPVTPGETR PA YDGYEAS KQIIIDAESA LNEWTLDSAQ VKPTLSFKQD YEDFEPEEGE EEEEEDGQGR WSEDEQEDGQ IEEPEQEEEG AVE EHEQVG HQQSESLPQQ TTA UniProtKB: Sphingolipid long chain base-responsive protein PIL1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: 20mM HEPES, pH 7.4, 150mM KoAc, 2mM MgAc |
|---|---|
| Grid | Model: EMS Lacey Carbon / Support film - Material: CARBON / Support film - topology: LACEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 291 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 5.33 Å Applied symmetry - Helical parameters - Δ&Phi: 133.61 ° Applied symmetry - Helical parameters - Axial symmetry: D1 (2x1 fold dihedral) Resolution.type: BY AUTHOR / Resolution: 3.85 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 56475 |
|---|---|
| Startup model | Type of model: NONE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)