-Search query
-Search result
Showing all 41 items for (author: hsieh & ky)
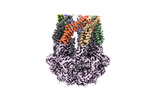
EMDB-36965: 
CryoEM structure of LonC protease hepatmer, apo state
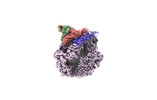
EMDB-36966: 
CryoEM structure of LonC protease open hexamer, apo state
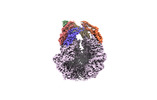
EMDB-36967: 
CryoEM of LonC open pentamer, apo state
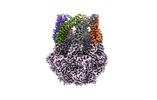
EMDB-36968: 
CryoEM structure of LonC heptamer in presence of AGS
EMDB-36969: 
CryoEM structure of LonC protease hexamer in presence of AGS
EMDB-36970: 
CryoEM structure of LonC protease open pentamer in presence of AGS
EMDB-36971: 
CryoEM structure of LonC S582A hepatmer with Lysozyme
EMDB-36972: 
CryoEM structure of LonC S582A hexamer with Lysozyme
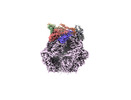
EMDB-36973: 
CryoEM structure of LonC protease S582A open hexamer with lysozyme
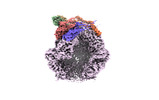
EMDB-36974: 
CryoEM structure of LonC protease S582A open pentamer with lysozyme
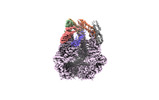
EMDB-36975: 
CryoEM structure of LonC protease open Hexamer, AGS
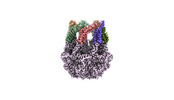
EMDB-36976: 
CryoEM structure of LonC protease hepatmer with Bortezomib
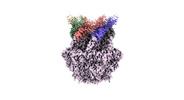
EMDB-36977: 
CryoEM structure of LonC protease hexamer with Bortezomib

EMDB-36978: 
CryoEM map of LonC protease open hexamer with Bortezomib

EMDB-36979: 
CryoEM map of LonC protease heptamer (heated at 343K)

EMDB-34000: 
Open-spiral pentamer of the substrate-free Lon protease with a Y224S mutation

EMDB-34001: 
Spiral hexamer of the substrate-free Lon protease with a Y224S mutation

EMDB-34002: 
Spiral pentamer of the substrate-free Lon protease with a S678A mutation

EMDB-34003: 
Close-ring hexamer of the substrate-bound Lon protease with an S678A mutation

EMDB-34004: 
Spiral hexamer of the substrate-free Lon protease with an S678A mutation

EMDB-34005: 
Open-spiral pentamer of the substrate-free Lon protease with Y397A and S678A mutations

EMDB-34006: 
Spiral hexamer of the substrate-free Lon protease with Y397A and S678A mutations

EMDB-34107: 
MtaLon-Apo for the spiral oligomers of trimer
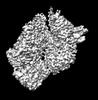
EMDB-34108: 
MtaLon-Apo for the spiral oligomers of tetramer

EMDB-34109: 
MtaLon-Apo for the spiral oligomers of pentamer

EMDB-34110: 
MtaLon-Apo for the spiral oligomers of hexamer
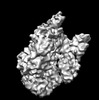
EMDB-34111: 
MtaLon-ADP for the spiral oligomers of trimer
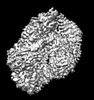
EMDB-34112: 
MtaLon-ADP for the spiral oligomers of tetramer

EMDB-34113: 
MtaLon-ADP for the spiral oligomers of pentamer

EMDB-34114: 
MtaLon-ADP for the spiral oligomers of hexamer

EMDB-34116: 
Spiral pentamer of the substrate-free Lon protease with an M217A mutation

EMDB-34117: 
Spiral hexamer of the substrate-free Lon protease with an M217A mutation

EMDB-36865: 
Spiral pentameric form of the substrate-free Lon protease with a Y224S mutation in the presence of the N-terminal-truncated monomeric mutant bearing an E613K mutation

EMDB-36866: 
Spiral hexameric form of the substrate-free Lon protease with a Y224S mutation in the presence of the N-terminal-truncated monomeric mutant bearing an E613K mutation

EMDB-36867: 
The "5+1" heteromeric structure of Lon protease consisting of a spiral pentamer with Y224S mutation and an N-terminal-truncated monomeric E613K mutant

EMDB-31589: 
Processive cleavage of substrate at individual proteolytic active sites of the Lon proteasecomplex (conformation 1)

EMDB-31590: 
Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 2)

EMDB-31607: 
Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 3)

EMDB-31534: 
A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 1)

EMDB-31535: 
A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 2)

EMDB-21870: 
Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
