-Search query
-Search result
Showing 1 - 50 of 261 items for (author: howard & g)

EMDB-43011: 
Phosphorylated, ATP-bound, E1371Q human cystic fibrosis transmembrane conductance regulator (E1371Q-CFTR)

EMDB-43014: 
Phosphorylated, ATP-bound, inhibitor 172-bound E1371Q human cystic fibrosis transmembrane conductance regulator

EMDB-42074: 
Representative tomogram of Enterococcus faecium WT Com15

EMDB-42086: 
Representative tomogram of Enterococcus faecium SagA complementation strain

EMDB-42087: 
Representative tomogram of Enterococcus faecium SagA deletion strain

EMDB-41248: 
Structure of AT118-H Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor

EMDB-41249: 
Structure of AT118-L Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor and Losartan

PDB-8th3: 
Structure of AT118-H Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor

PDB-8th4: 
Structure of AT118-L Nanobody Antagonist in Complex with the Angiotensin II Type I Receptor and Losartan
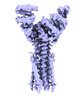
EMDB-50106: 
Artificial membrane protein TMHC4_R (ROCKET)
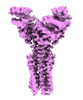
EMDB-50107: 
Artificial membrane protein TMHC4_R (ROCKET) mutant R9A/K10A/R13A

EMDB-42681: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the upper strand

EMDB-42682: 
The structure of the native cardiac thin filament troponin core in Ca2+-free tilted state from the upper strand

EMDB-42683: 
The structure of the native cardiac thin filament troponin core in Ca2+-free rotated state from the upper strand

EMDB-42800: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the lower strand

EMDB-42833: 
The structure of the native cardiac thin filament troponin core in Ca2+-free rotated state from the lower strand

EMDB-42835: 
The structure of the native cardiac thin filament troponin core in Ca2+-free tilted state from the lower strand

EMDB-42846: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state from the upper strand

EMDB-42847: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound partially activated state from the upper strand

EMDB-42849: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state 1 from the lower strand

EMDB-42856: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state 2 from the lower strand

EMDB-42858: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound partially activated state from the lower strand

EMDB-42874: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the upper strand activated by the C1-domain of cardiac myosin binding protein C

PDB-8uww: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the upper strand

PDB-8uwx: 
The structure of the native cardiac thin filament troponin core in Ca2+-free tilted state from the upper strand

PDB-8uwy: 
The structure of the native cardiac thin filament troponin core in Ca2+-free rotated state from the upper strand

PDB-8uyd: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the lower strand

PDB-8uz5: 
The structure of the native cardiac thin filament troponin core in Ca2+-free rotated state from the lower strand

PDB-8uz6: 
The structure of the native cardiac thin filament troponin core in Ca2+-free tilted state from the lower strand

PDB-8uzx: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state from the upper strand

PDB-8uzy: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound partially activated state from the upper strand

PDB-8v01: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state 1 from the lower strand

PDB-8v0i: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state 2 from the lower strand

PDB-8v0k: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound partially activated state from the lower strand

PDB-8v0y: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the upper strand activated by the C1-domain of cardiac myosin binding protein C
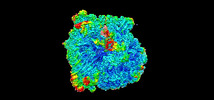
EMDB-42455: 
Candidatus Methanomethylophilus alvus tRNAPyl in A-site of ribosome

PDB-8upt: 
Candidatus Methanomethylophilus alvus tRNAPyl in A-site of ribosome

EMDB-29281: 
Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2

EMDB-29282: 
Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53

PDB-8flk: 
Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2

PDB-8flm: 
Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53

EMDB-28209: 
Apo rat TRPV2 in nanodiscs, state 1

EMDB-28210: 
Apo rat TRPV2 in nanodiscs, state 2

EMDB-28211: 
Apo rat TRPV2 in nanodiscs, state 3

EMDB-28212: 
rat TRPV2 in nanodiscs in the presence of weak acid at pH 5

PDB-8ekp: 
Apo rat TRPV2 in nanodiscs, state 1

PDB-8ekq: 
Apo rat TRPV2 in nanodiscs, state 2

PDB-8ekr: 
Apo rat TRPV2 in nanodiscs, state 3

PDB-8eks: 
rat TRPV2 in nanodiscs in the presence of weak acid at pH 5

EMDB-15649: 
Pentameric ligand-gated ion channel GLIC with bound lipids
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
