[English] 日本語
 Yorodumi
Yorodumi- EMDB-42682: The structure of the native cardiac thin filament troponin core i... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of the native cardiac thin filament troponin core in Ca2+-free tilted state from the upper strand | |||||||||
 Map data Map data | map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | thin filament / troponin / tropomyosin / cryo-EM / muscle structure / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationStriated Muscle Contraction / regulation of muscle filament sliding speed / troponin T binding / cardiac Troponin complex / troponin complex / actin-myosin filament sliding / regulation of muscle contraction / transition between fast and slow fiber / ventricular cardiac muscle tissue morphogenesis / heart contraction ...Striated Muscle Contraction / regulation of muscle filament sliding speed / troponin T binding / cardiac Troponin complex / troponin complex / actin-myosin filament sliding / regulation of muscle contraction / transition between fast and slow fiber / ventricular cardiac muscle tissue morphogenesis / heart contraction / myosin binding / troponin I binding / mesenchyme migration / skeletal muscle contraction / cardiac muscle contraction / actin filament organization / sarcomere / actin filament / filopodium / calcium-dependent protein binding / actin filament binding / actin cytoskeleton / lamellipodium / actin binding / cell body / protein heterodimerization activity / calcium ion binding / positive regulation of gene expression / protein homodimerization activity / ATP binding / identical protein binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
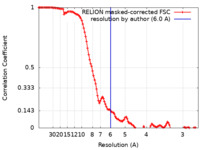
| Method | single particle reconstruction / cryo EM / Resolution: 6.0 Å | |||||||||
 Authors Authors | Galkin VE / Risi CM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2024 Journal: J Mol Biol / Year: 2024Title: Troponin Structural Dynamics in the Native Cardiac Thin Filament Revealed by Cryo Electron Microscopy. Authors: Cristina M Risi / Betty Belknap / Jennifer Atherton / Isabella Leite Coscarella / Howard D White / P Bryant Chase / Jose R Pinto / Vitold E Galkin /  Abstract: Cardiac muscle contraction occurs due to repetitive interactions between myosin thick and actin thin filaments (TF) regulated by Ca levels, active cross-bridges, and cardiac myosin-binding protein C ...Cardiac muscle contraction occurs due to repetitive interactions between myosin thick and actin thin filaments (TF) regulated by Ca levels, active cross-bridges, and cardiac myosin-binding protein C (cMyBP-C). The cardiac TF (cTF) has two nonequivalent strands, each comprised of actin, tropomyosin (Tm), and troponin (Tn). Tn shifts Tm away from myosin-binding sites on actin at elevated Ca levels to allow formation of force-producing actomyosin cross-bridges. The Tn complex is comprised of three distinct polypeptides - Ca-binding TnC, inhibitory TnI, and Tm-binding TnT. The molecular mechanism of their collective action is unresolved due to lack of comprehensive structural information on Tn region of cTF. C1 domain of cMyBP-C activates cTF in the absence of Ca to the same extent as rigor myosin. Here we used cryo-EM of native cTFs to show that cTF Tn core adopts multiple structural conformations at high and low Ca levels and that the two strands are structurally distinct. At high Ca levels, cTF is not entirely activated by Ca but exists in either partially or fully activated state. Complete dissociation of TnI C-terminus is required for full activation. In presence of cMyBP-C C1 domain, Tn core adopts a fully activated conformation, even in absence of Ca. Our data provide a structural description for the requirement of myosin to fully activate cTFs and explain increased affinity of TnC to Ca in presence of active cross-bridges. We suggest that allosteric coupling between Tn subunits and Tm is required to control actomyosin interactions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42682.map.gz emd_42682.map.gz | 2.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42682-v30.xml emd-42682-v30.xml emd-42682.xml emd-42682.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42682_fsc.xml emd_42682_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_42682.png emd_42682.png | 83.6 KB | ||
| Filedesc metadata |  emd-42682.cif.gz emd-42682.cif.gz | 6.6 KB | ||
| Others |  emd_42682_half_map_1.map.gz emd_42682_half_map_1.map.gz emd_42682_half_map_2.map.gz emd_42682_half_map_2.map.gz | 102.3 MB 102.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42682 http://ftp.pdbj.org/pub/emdb/structures/EMD-42682 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42682 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42682 | HTTPS FTP |
-Validation report
| Summary document |  emd_42682_validation.pdf.gz emd_42682_validation.pdf.gz | 765.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42682_full_validation.pdf.gz emd_42682_full_validation.pdf.gz | 765.3 KB | Display | |
| Data in XML |  emd_42682_validation.xml.gz emd_42682_validation.xml.gz | 19.2 KB | Display | |
| Data in CIF |  emd_42682_validation.cif.gz emd_42682_validation.cif.gz | 25.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42682 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42682 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42682 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42682 | HTTPS FTP |
-Related structure data
| Related structure data |  8uwxMC  8uwwC  8uwyC  8uydC  8uz5C  8uz6C  8uzxC  8uzyC  8v01C  8v0iC  8v0kC  8v0yC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42682.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42682.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.356 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half1
| File | emd_42682_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half2
| File | emd_42682_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : thin filament troponin core complex
| Entire | Name: thin filament troponin core complex |
|---|---|
| Components |
|
-Supramolecule #1: thin filament troponin core complex
| Supramolecule | Name: thin filament troponin core complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Actin, alpha cardiac muscle 1
| Macromolecule | Name: Actin, alpha cardiac muscle 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.064891 KDa |
| Sequence | String: MCDDEETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIEHGIITN WDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLDSGD G VTHNVPIY ...String: MCDDEETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIEHGIITN WDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLDSGD G VTHNVPIY EGYALPHAIM RLDLAGRDLT DYLMKILTER GYSFVTTAER EIVRDIKEKL CYVALDFENE MATAASSSSL EK SYELPDG QVITIGNERF RCPETLFQPS FIGMESAGIH ETTYNSIMKC DIDIRKDLYA NNVLSGGTTM YPGIADRMQK EIT ALAPST MKIKIIAPPE RKYSVWIGGS ILASLSTFQQ MWISKQEYDE AGPSIVHRKC F UniProtKB: Actin alpha cardiac muscle 1 |
-Macromolecule #2: Troponin C, slow skeletal and cardiac muscles
| Macromolecule | Name: Troponin C, slow skeletal and cardiac muscles / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.433508 KDa |
| Sequence | String: MDDIYKAAVE QLTEEQKNEF KAAFDIFVLG AEDGCISTKE LGKVMRMLGQ NPTPEELQEM IDEVDEDGSG TVDFDEFLVM MVRCMKDDS KGKSEEELSD LFRMFDKNAD GYIDLEELKI MLQATGETIT EDDIEELMKD GDKNNDGRID YDEFLEFMKG V E UniProtKB: Troponin C, slow skeletal and cardiac muscles |
-Macromolecule #3: Troponin I, cardiac muscle
| Macromolecule | Name: Troponin I, cardiac muscle / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.09268 KDa |
| Sequence | String: MADRSGDAAG DSRPAPAPVR RRSSANYRAY ATEPHAKKKS KISASRKLQL KTLMLQIAKQ ELEREAEERR GEKGRALSTR CQPLELAGL SFAELQDLCR QLHARVDKVD EERYDVEAKV TKNITEIADL NQKIFDLRGK FKRPTLRRVR ISADAMMQAL L GARAKETL ...String: MADRSGDAAG DSRPAPAPVR RRSSANYRAY ATEPHAKKKS KISASRKLQL KTLMLQIAKQ ELEREAEERR GEKGRALSTR CQPLELAGL SFAELQDLCR QLHARVDKVD EERYDVEAKV TKNITEIADL NQKIFDLRGK FKRPTLRRVR ISADAMMQAL L GARAKETL DLRAHLKQVK KEDTEKENRE VGDWRKNIDA LSGMEGRKKK FEG UniProtKB: Troponin I, cardiac muscle |
-Macromolecule #4: Troponin T2, cardiac type
| Macromolecule | Name: Troponin T2, cardiac type / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.941738 KDa |
| Sequence | String: MSDVEETVDE YEEEQEEGAA EEQEEAVEEE AGGEAEAEEA NAEEAGQEED GREAEDGPME ESKPKPRLFM PNLVPPKIPD GERVDFDDI HRKRMKDLNE LQTLIEAHFE NRKKEEELVS LKDRIEKRRA ERAEQQRIRT EREKERQTRL AEERARREEE E NRRKAEDE ...String: MSDVEETVDE YEEEQEEGAA EEQEEAVEEE AGGEAEAEEA NAEEAGQEED GREAEDGPME ESKPKPRLFM PNLVPPKIPD GERVDFDDI HRKRMKDLNE LQTLIEAHFE NRKKEEELVS LKDRIEKRRA ERAEQQRIRT EREKERQTRL AEERARREEE E NRRKAEDE ARKKKALSNM MHFGGYIQKQ AQTERKSGKR QTEREKKKKI LAERRKVLAI DHLNEDQLRE KAKELWQSIY NL EAEKFDL QEKFKQQKYE INVLRNRIND NQKVSKTRGK AKVTGRWK UniProtKB: Troponin T, cardiac muscle |
-Macromolecule #5: Tropomyosin alpha-1 chain
| Macromolecule | Name: Tropomyosin alpha-1 chain / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.762656 KDa |
| Sequence | String: MDAIKKKMQM LKLDKENALD RAEQAEADKK AAEDRSKRLE DELVSLQKKL KATEDELDKY SEAPKDAQEK LELAEKKATD AEADVASLN RRIQLVEEEL DRAQERLATA LQKLEEAEKA ADESERGMKV IESRAQKDEE KMEIQEIQLK EAKHIAEDAD R KYEEVARK ...String: MDAIKKKMQM LKLDKENALD RAEQAEADKK AAEDRSKRLE DELVSLQKKL KATEDELDKY SEAPKDAQEK LELAEKKATD AEADVASLN RRIQLVEEEL DRAQERLATA LQKLEEAEKA ADESERGMKV IESRAQKDEE KMEIQEIQLK EAKHIAEDAD R KYEEVARK LVIIESDLER AEERAELSEG KCAELEEELK TVTNNLKSLE AQAEKYSQKE DKYEEEIKVL SDKLKEAETR AE FAERSVT KLEKSIDDLE DELYAQKLKY KAISEELDHA LNDMTSI UniProtKB: Tropomyosin alpha-1 chain |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 24133 / Average electron dose: 34.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||||
| Output model |  PDB-8uwx: |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)