-Search query
-Search result
Showing all 47 items for (author: halfmann & p)

EMDB-48548: 
SARS-CoV-2 S2 monomer in complex with R125-61 Fab
Method: single particle / : Park S, Bangaru B, Ward AB

EMDB-48549: 
SARS-CoV-2 S2 monomer in complex with NICA01B-1113 Fab
Method: single particle / : Park S, Bangaru B, Ward AB

EMDB-48550: 
SARS-CoV-2 S2 monomer in complex with NICA01A-1401 Fab
Method: single particle / : Park S, Bangaru B, Ward AB

PDB-9mr1: 
SARS-CoV-2 S2 monomer in complex with R125-61 Fab
Method: single particle / : Park S, Bangaru B, Ward AB

PDB-9mr2: 
SARS-CoV-2 S2 monomer in complex with NICA01A-1401 Fab
Method: single particle / : Park S, Bangaru B, Ward AB

EMDB-44747: 
A broadly-neutralizing antibody against Ebolavirus glycoprotein that can potentiate the breadth and neutralization potency of other anti-glycoprotein antibodies
Method: single particle / : Donnellan FR, Rayaprolu V, Rijal P, O'Dowd V, Parvate A, Callaway H, Hariharan C, Parekh D, Hui S, Shaffer K, Hastie K, Shimanski L, Muller-Krauter H, Stecker T, Balaram A, Halfmann P, Saphire EO, Lightwood DJ, Townsend AR, Draper SJ

PDB-9bop: 
A broadly-neutralizing antibody against Ebolavirus glycoprotein that can potentiate the breadth and neutralization potency of other anti-glycoprotein antibodies
Method: single particle / : Donnellan FR, Rayaprolu V, Rijal P, O'Dowd V, Parvate A, Callaway H, Hariharan C, Parekh D, Hui S, Shaffer K, Hastie K, Shimanski L, Muller-Krauter H, Stecker T, Balaram A, Halfmann P, Saphire EO, Lightwood DJ, Townsend AR, Draper SJ

EMDB-46892: 
Structure of SARS-CoV-2 spike in complex with antibody Fab COVIC-154
Method: single particle / : Yu X, Saphire EO

PDB-9dhy: 
Structure of SARS-CoV-2 spike in complex with antibody Fab COVIC-154
Method: single particle / : Yu X, Saphire EO

EMDB-45833: 
KZ52 Fab fragment bound to Ebola GP, subtomogram average from a tight mask
Method: subtomogram averaging / : Ke Z, Saphire EO, Briggs JAG

EMDB-45834: 
KZ52 Fab fragment bound to Ebola GP, subtomogram average from a cylinder mask
Method: subtomogram averaging / : Ke Z, Saphire EO, Briggs JAG

EMDB-45835: 
3A6 Fab fragment bound to Ebola GP, subtomogram average from a cylinder mask
Method: subtomogram averaging / : Ke Z, Saphire EO, Briggs JAG

EMDB-45836: 
3A6 and KZ52 Fab fragments bound to Ebola GP, subtomogram average from a tight mask
Method: subtomogram averaging / : Ke Z, Saphire EO, Briggs JAG

EMDB-45837: 
3A6 and KZ52 Fab fragments bound to Ebola GP, subtomogram average from a cylinder mask
Method: subtomogram averaging / : Ke Z, Saphire EO, Briggs JAG

EMDB-27112: 
S728-1157 IgG in complex with SARS-CoV-2-6P-Mut7 Spike protein (global refinement)
Method: single particle / : Ozorowski G, Torres JL, Turner HL, Ward AB

EMDB-27113: 
S728-1157 IgG in complex with SARS-CoV-2-6P-Mut7 Spike protein (focused refinement)
Method: single particle / : Ozorowski G, Torres JL, Turner HL, Ward AB

PDB-8d0z: 
S728-1157 IgG in complex with SARS-CoV-2-6P-Mut7 Spike protein (focused refinement)
Method: single particle / : Ozorowski G, Torres JL, Ward AB

EMDB-25471: 
Structure of EBOV GP lacking the mucin-like domain with 1C11 scFv and 1C3 Fab bound
Method: single particle / : Milligan JC, Yu X

PDB-7swd: 
Structure of EBOV GP lacking the mucin-like domain with 1C11 scFv and 1C3 Fab bound
Method: single particle / : Milligan JC, Yu X, Saphire EO

EMDB-9332: 
Helical assembly of the CARD9 CARD
Method: helical / : Holliday MJ, Rohou A

PDB-6n2p: 
Helical assembly of the CARD9 CARD
Method: helical / : Holliday MJ, Rohou A, Arthur CP, Dueber EC, Fairbrother WJ
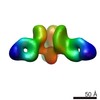
EMDB-7908: 
VIC170 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7909: 
VIC169 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
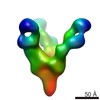
EMDB-7910: 
VIC167 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
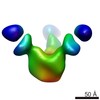
EMDB-7911: 
VIC166 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
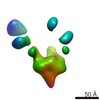
EMDB-7912: 
VIC165 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7914: 
VIC1164 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
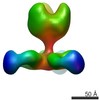
EMDB-7915: 
VIC163 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7926: 
VIC82 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
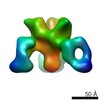
EMDB-7927: 
VIC43 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7928: 
VIC32 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7929: 
VIC26 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7930: 
VIC16 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7931: 
VIC15 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7932: 
VIC12 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
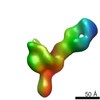
EMDB-7933: 
VIC11 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7922: 
VIC133 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7923: 
VIC101 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7924: 
VIC94 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7925: 
VIC91 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7934: 
VIC130 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
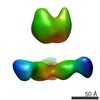
EMDB-7916: 
VIC161 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7917: 
VIC160 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
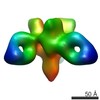
EMDB-7918: 
VIC158 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
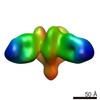
EMDB-7919: 
VIC157 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7920: 
VIC142 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
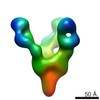
EMDB-7921: 
VIC139 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
