+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7934 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | VIC130 Fab in complex with Ebola virus GP | |||||||||
 Map data Map data | VIC130 antibody Fab in complex with Ebola virus GP | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 26.0 Å | |||||||||
 Authors Authors | Turner H / Murin CD / Pallesen J / Ward AB | |||||||||
 Citation Citation |  Journal: Cell / Year: 2018 Journal: Cell / Year: 2018Title: Systematic Analysis of Monoclonal Antibodies against Ebola Virus GP Defines Features that Contribute to Protection. Authors: Erica Ollmann Saphire / Sharon L Schendel / Marnie L Fusco / Karthik Gangavarapu / Bronwyn M Gunn / Anna Z Wec / Peter J Halfmann / Jennifer M Brannan / Andrew S Herbert / Xiangguo Qiu / ...Authors: Erica Ollmann Saphire / Sharon L Schendel / Marnie L Fusco / Karthik Gangavarapu / Bronwyn M Gunn / Anna Z Wec / Peter J Halfmann / Jennifer M Brannan / Andrew S Herbert / Xiangguo Qiu / Kshitij Wagh / Shihua He / Elena E Giorgi / James Theiler / Kathleen B J Pommert / Tyler B Krause / Hannah L Turner / Charles D Murin / Jesper Pallesen / Edgar Davidson / Rafi Ahmed / M Javad Aman / Alexander Bukreyev / Dennis R Burton / James E Crowe / Carl W Davis / George Georgiou / Florian Krammer / Christos A Kyratsous / Jonathan R Lai / Cory Nykiforuk / Michael H Pauly / Pramila Rijal / Ayato Takada / Alain R Townsend / Viktor Volchkov / Laura M Walker / Cheng-I Wang / Larry Zeitlin / Benjamin J Doranz / Andrew B Ward / Bette Korber / Gary P Kobinger / Kristian G Andersen / Yoshihiro Kawaoka / Galit Alter / Kartik Chandran / John M Dye / /       Abstract: Antibodies are promising post-exposure therapies against emerging viruses, but which antibody features and in vitro assays best forecast protection are unclear. Our international consortium ...Antibodies are promising post-exposure therapies against emerging viruses, but which antibody features and in vitro assays best forecast protection are unclear. Our international consortium systematically evaluated antibodies against Ebola virus (EBOV) using multidisciplinary assays. For each antibody, we evaluated epitopes recognized on the viral surface glycoprotein (GP) and secreted glycoprotein (sGP), readouts of multiple neutralization assays, fraction of virions left un-neutralized, glycan structures, phagocytic and natural killer cell functions elicited, and in vivo protection in a mouse challenge model. Neutralization and induction of multiple immune effector functions (IEFs) correlated most strongly with protection. Neutralization predominantly occurred via epitopes maintained on endosomally cleaved GP, whereas maximal IEF mapped to epitopes farthest from the viral membrane. Unexpectedly, sGP cross-reactivity did not significantly influence in vivo protection. This comprehensive dataset provides a rubric to evaluate novel antibodies and vaccine responses and a roadmap for therapeutic development for EBOV and related viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7934.map.gz emd_7934.map.gz | 950.1 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7934-v30.xml emd-7934-v30.xml emd-7934.xml emd-7934.xml | 13.2 KB 13.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7934.png emd_7934.png | 24.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7934 http://ftp.pdbj.org/pub/emdb/structures/EMD-7934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7934 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7934 | HTTPS FTP |
-Related structure data
| Related structure data | 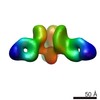 7908C  7909C 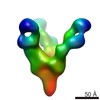 7910C  7911C  7912C  7914C 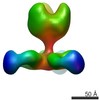 7915C  7916C  7917C  7918C  7919C  7920C  7921C  7922C  7923C  7924C  7925C  7926C  7927C  7928C  7929C  7930C  7931C  7932C 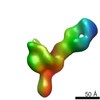 7933C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7934.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7934.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | VIC130 antibody Fab in complex with Ebola virus GP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
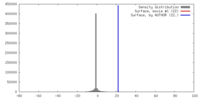
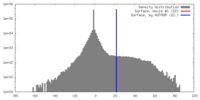
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of VIC130 Fab bound to Ebola virus GP
| Entire | Name: Complex of VIC130 Fab bound to Ebola virus GP |
|---|---|
| Components |
|
-Supramolecule #1: Complex of VIC130 Fab bound to Ebola virus GP
| Supramolecule | Name: Complex of VIC130 Fab bound to Ebola virus GP / type: complex / ID: 1 / Parent: 0 Details: Fab fragment generated by proteolytic cleavage of IgG antibody with papain. |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: Solution was made from 10X concentration. | |||||||||
| Staining | Type: NEGATIVE / Material: uranyl formate / Details: Stained using 2% UF on carbon coated grids. | |||||||||
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER | |||||||||
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 97 / Average exposure time: 0.5 sec. / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus max: 0.001 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: Common lines model. |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 26.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN2 (ver. 2.1) / Number images used: 10551 |
| Initial angle assignment | Type: COMMON LINE / Software - Name: EMAN2 |
| Final angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: EMAN2 |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)