-Search query
-Search result
Showing 1 - 50 of 115 items for (author: hale & wa)

EMDB-43931: 
CryoEM structure of activated CRAF/MEK/14-3-3 complex with NST-628

EMDB-43932: 
Activated CRAF/MEK heterotetramer from focused refinement of CRAF/MEK/14-3-3 complex

PDB-9axa: 
CryoEM structure of activated CRAF/MEK/14-3-3 complex with NST-628

PDB-9axc: 
Activated CRAF/MEK heterotetramer from focused refinement of CRAF/MEK/14-3-3 complex

EMDB-42601: 
CryoEM structure of Kappa Opioid Receptor bound to a semi-peptide and Gi1

EMDB-29026: 
CryoEM structure of Kappa Opioid Receptor bound to a semi-peptide and Gi1

PDB-8feg: 
CryoEM structure of Kappa Opioid Receptor bound to a semi-peptide and Gi1

EMDB-17756: 
Structure of the murine trace amine-associated receptor TAAR7f bound to N,N-dimethylcyclohexylamine (DMCH) in complex with mini-Gs trimeric G protein

PDB-8pm2: 
Structure of the murine trace amine-associated receptor TAAR7f bound to N,N-dimethylcyclohexylamine (DMCH) in complex with mini-Gs trimeric G protein

EMDB-15786: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli (Apo form)

EMDB-15787: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE

EMDB-15788: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE (C387S mutant)

EMDB-15789: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Lyso-PE

EMDB-15790: 
Cryo-EM structure apolipoprotein N-acyltransferase Lnt from E.coli in complex with FP3

EMDB-15791: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Pam3

PDB-8b0k: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli (Apo form)

PDB-8b0l: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE

PDB-8b0m: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE (C387S mutant)

PDB-8b0n: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Lyso-PE

PDB-8b0o: 
Cryo-EM structure apolipoprotein N-acyltransferase Lnt from E.coli in complex with FP3

PDB-8b0p: 
Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Pam3

EMDB-27692: 
LM18/Nb136 bispecific tetra-nanobody immunoglobulin in complex with SARS-CoV-2-6P-Mut7 S protein (focused refinement)

EMDB-27693: 
LM18/Nb136 bispecific tetra-nanobody immunoglobulin in complex with SARS-CoV-2-6P-Mut7 S protein (global refinement)

PDB-8dt8: 
LM18/Nb136 bispecific tetra-nanobody immunoglobulin in complex with SARS-CoV-2-6P-Mut7 S protein (focused refinement)

EMDB-29044: 
Structure of Zanidatamab bound to HER2
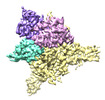
EMDB-27898: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA
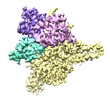
EMDB-27899: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA

PDB-8e4y: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with 2-oxohexadecyl-CoA

PDB-8e50: 
Cryo-EM structure of human glycerol-3-phosphate acyltransferase 1 (GPAT1) in complex with CoA and palmitoyl-LPA

EMDB-13485: 
Cryo-EM structure of Bestrhodopsin (rhodopsin-rhodopsin-bestrophin) complex

PDB-7pl9: 
Cryo-EM structure of Bestrhodopsin (rhodopsin-rhodopsin-bestrophin) complex

EMDB-25076: 
LPHN3 (ADGRL3) 7TM domain bound to tethered agonist in complex with G protein heterotrimer

EMDB-25077: 
GPR56 (ADGRG1) 7TM domain bound to tethered agonist in complex with G protein heterotrimer

PDB-7sf7: 
LPHN3 (ADGRL3) 7TM domain bound to tethered agonist in complex with G protein heterotrimer

PDB-7sf8: 
GPR56 (ADGRG1) 7TM domain bound to tethered agonist in complex with G protein heterotrimer

EMDB-25634: 
Negative stain map of monoclonal Fab 047-09 4F04 binding the anchor epitope of H1 HA

EMDB-25635: 
Negative stain map of monoclonal Fab 241 IgA 2F04 binding the anchor epitope of H1 HA

EMDB-25636: 
Negative stain map of polyclonal Fab 236.7 binding the anchor and esterase epitopes of H1 HA

EMDB-25637: 
Negative stain map of polyclonal Fab 236.7 binding the RBS epitope of H1 HA

EMDB-25638: 
Negative stain map of polyclonal Fab 236.14 binding an epitope on the top of the head of H1 HA
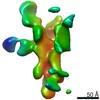
EMDB-25639: 
Negative stain map of polyclonal Fab 236.14 binding the esterase epitope of H1 HA

EMDB-25640: 
Negative stain map of polycolonal Fab 236.14 binding the RBS epitope of H1 HA

EMDB-25641: 
Negative stain map of polyclonal Fab 236.14 binding the anchor epitope of H1 HA
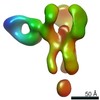
EMDB-25642: 
Negative stain map of polyclonal Fab 241.7 binding the esterase epitope of H1 HA
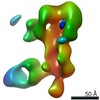
EMDB-25643: 
Negative stain map of polyclonal Fab 241.14 binding the anchor epitope of H1 HA
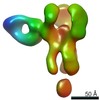
EMDB-25644: 
Negative stain map of polyclonal Fab 241.14 binding the esterase epitope of H1 HA

EMDB-25645: 
Negative stain map of polyclonal Fab 241.14 binding an epitope on the top of the head of H1 HA

EMDB-25646: 
Negative stain map of polyclonal Fab 241.14 binding the RBS epitope of H1 HA

EMDB-25655: 
CryoEM map of anchor 222-1C06 Fab and lateral patch 2B05 Fab binding H1 HA

PDB-7t3d: 
CryoEM map of anchor 222-1C06 Fab and lateral patch 2B05 Fab binding H1 HA
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
