[English] 日本語
 Yorodumi
Yorodumi- EMDB-15786: Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E.... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli (Apo form) | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Lnt / apolipoprotein N-acyltransferase / bacterial lipoprotein / transferase / cryo-EM | |||||||||
| Function / homology |  Function and homology information Function and homology informationapolipoprotein N-acyltransferase / N-acyltransferase activity / lipoprotein biosynthetic process / outer membrane-bounded periplasmic space / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
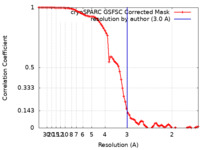
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Degtjarik O / Smithers L / Boland C / Caffrey M / Shalev Benami M | |||||||||
| Funding support |  Ireland, 2 items Ireland, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein -acyltransferase. Authors: Luke Smithers / Oksana Degtjarik / Dietmar Weichert / Chia-Ying Huang / Coilín Boland / Katherine Bowen / Abraham Oluwole / Corinne Lutomski / Carol V Robinson / Eoin M Scanlan / Meitian ...Authors: Luke Smithers / Oksana Degtjarik / Dietmar Weichert / Chia-Ying Huang / Coilín Boland / Katherine Bowen / Abraham Oluwole / Corinne Lutomski / Carol V Robinson / Eoin M Scanlan / Meitian Wang / Vincent Olieric / Moran Shalev-Benami / Martin Caffrey /     Abstract: Bacterial lipoproteins (BLPs) decorate the surface of membranes in the cell envelope. They function in membrane assembly and stability, as enzymes, and in transport. The final enzyme in the BLP ...Bacterial lipoproteins (BLPs) decorate the surface of membranes in the cell envelope. They function in membrane assembly and stability, as enzymes, and in transport. The final enzyme in the BLP synthesis pathway is the apolipoprotein -acyltransferase, Lnt, which is proposed to act by a ping-pong mechanism. Here, we use x-ray crystallography and cryo-electron microscopy to chart the structural changes undergone during the progress of the enzyme through the reaction. We identify a single active site that has evolved to bind, individually and sequentially, substrates that satisfy structural and chemical criteria to position reactive parts next to the catalytic triad for reaction. This study validates the ping-pong mechanism, explains the molecular bases for Lnt's substrate promiscuity, and should facilitate the design of antibiotics with minimal off-target effects. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15786.map.gz emd_15786.map.gz | 27.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15786-v30.xml emd-15786-v30.xml emd-15786.xml emd-15786.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15786_fsc.xml emd_15786_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_15786.png emd_15786.png | 36.3 KB | ||
| Filedesc metadata |  emd-15786.cif.gz emd-15786.cif.gz | 5.8 KB | ||
| Others |  emd_15786_additional_1.map.gz emd_15786_additional_1.map.gz emd_15786_half_map_1.map.gz emd_15786_half_map_1.map.gz emd_15786_half_map_2.map.gz emd_15786_half_map_2.map.gz | 26.4 MB 49 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15786 http://ftp.pdbj.org/pub/emdb/structures/EMD-15786 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15786 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15786 | HTTPS FTP |
-Related structure data
| Related structure data |  8b0kMC  8aq2C  8aq3C  8aq4C  8b0lC  8b0mC  8b0nC  8b0oC  8b0pC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15786.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15786.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.826 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: unsharpened map
| File | emd_15786_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map
| File | emd_15786_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map
| File | emd_15786_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Apolipoprotein N-acyltransferase
| Entire | Name: Apolipoprotein N-acyltransferase |
|---|---|
| Components |
|
-Supramolecule #1: Apolipoprotein N-acyltransferase
| Supramolecule | Name: Apolipoprotein N-acyltransferase / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Apolipoprotein N-acyltransferase
| Macromolecule | Name: Apolipoprotein N-acyltransferase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Transferases; Acyltransferases; Transferring groups other than aminoacyl groups |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 59.280766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MAFASLIERQ RIRLLLALLF GACGTLAFSP YDVWPAAIIS LMGLQALTFN RRPLQSAAIG FCWGFGLFG SGINWVYVSI ATFGGMPGPV NIFLVVLLAA YLSLYTGLFA GVLSRLWPKT TWLRVAIAAP ALWQVTEFLR G WVLTGFPW ...String: MGSSHHHHHH SSGLVPRGSH MAFASLIERQ RIRLLLALLF GACGTLAFSP YDVWPAAIIS LMGLQALTFN RRPLQSAAIG FCWGFGLFG SGINWVYVSI ATFGGMPGPV NIFLVVLLAA YLSLYTGLFA GVLSRLWPKT TWLRVAIAAP ALWQVTEFLR G WVLTGFPW LQFGYSQIDG PLKGLAPIMG VEAINFLLMM VSGLLALALV KRNWRPLVVA VVLFALPFPL RYIQWFTPQP EK TIQVSMV QGDIPQSLKW DEGQLLNTLK IYYNATAPLM GKSSLIIWPE SAITDLEINQ QPFLKALDGE LRDKGSSLVT GIV DARLNK QNRYDTYNTI ITLGKGAPYS YESADRYNKN HLVPFGEFVP LESILRPLAP FFDLPMSSFS RGPYIQPPLS ANGI ELTAA ICYEIILGEQ VRDNFRPDTD YLLTISNDAW FGKSIGPWQH FQMARMRALE LARPLLRSTN NGITAVIGPQ GEIQA MIPQ FTREVLTTNV TPTTGLTPYA RTGNWPLWVL TALFGFAAVL MSLRQRRK UniProtKB: Apolipoprotein N-acyltransferase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 14 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6 Component:
| ||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 33.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)